Xuewei Cao
Multi-View Variational Autoencoder for Missing Value Imputation in Untargeted Metabolomics
Oct 12, 2023


Abstract:Background: Missing data is a common challenge in mass spectrometry-based metabolomics, which can lead to biased and incomplete analyses. The integration of whole-genome sequencing (WGS) data with metabolomics data has emerged as a promising approach to enhance the accuracy of data imputation in metabolomics studies. Method: In this study, we propose a novel method that leverages the information from WGS data and reference metabolites to impute unknown metabolites. Our approach utilizes a multi-view variational autoencoder to jointly model the burden score, polygenetic risk score (PGS), and linkage disequilibrium (LD) pruned single nucleotide polymorphisms (SNPs) for feature extraction and missing metabolomics data imputation. By learning the latent representations of both omics data, our method can effectively impute missing metabolomics values based on genomic information. Results: We evaluate the performance of our method on empirical metabolomics datasets with missing values and demonstrate its superiority compared to conventional imputation techniques. Using 35 template metabolites derived burden scores, PGS and LD-pruned SNPs, the proposed methods achieved r2-scores > 0.01 for 71.55% of metabolites. Conclusion: The integration of WGS data in metabolomics imputation not only improves data completeness but also enhances downstream analyses, paving the way for more comprehensive and accurate investigations of metabolic pathways and disease associations. Our findings offer valuable insights into the potential benefits of utilizing WGS data for metabolomics data imputation and underscore the importance of leveraging multi-modal data integration in precision medicine research.
CLCLSA: Cross-omics Linked embedding with Contrastive Learning and Self Attention for multi-omics integration with incomplete multi-omics data
Apr 12, 2023



Abstract:Integration of heterogeneous and high-dimensional multi-omics data is becoming increasingly important in understanding genetic data. Each omics technique only provides a limited view of the underlying biological process and integrating heterogeneous omics layers simultaneously would lead to a more comprehensive and detailed understanding of diseases and phenotypes. However, one obstacle faced when performing multi-omics data integration is the existence of unpaired multi-omics data due to instrument sensitivity and cost. Studies may fail if certain aspects of the subjects are missing or incomplete. In this paper, we propose a deep learning method for multi-omics integration with incomplete data by Cross-omics Linked unified embedding with Contrastive Learning and Self Attention (CLCLSA). Utilizing complete multi-omics data as supervision, the model employs cross-omics autoencoders to learn the feature representation across different types of biological data. The multi-omics contrastive learning, which is used to maximize the mutual information between different types of omics, is employed before latent feature concatenation. In addition, the feature-level self-attention and omics-level self-attention are employed to dynamically identify the most informative features for multi-omics data integration. Extensive experiments were conducted on four public multi-omics datasets. The experimental results indicated that the proposed CLCLSA outperformed the state-of-the-art approaches for multi-omics data classification using incomplete multi-omics data.
Hip Fracture Prediction using the First Principal Component Derived from FEA-Computed Fracture Loads
Oct 03, 2022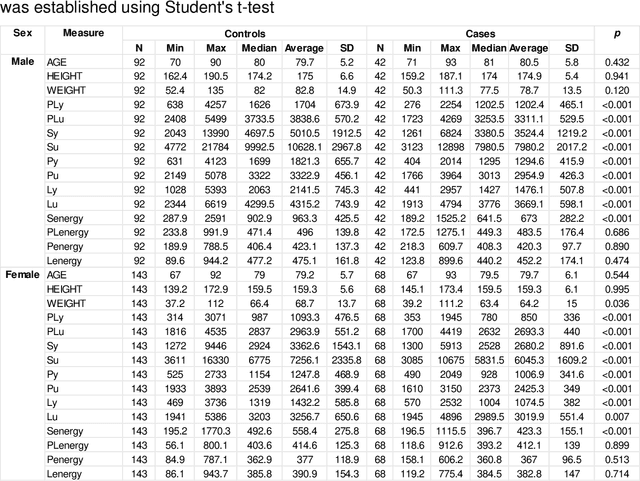
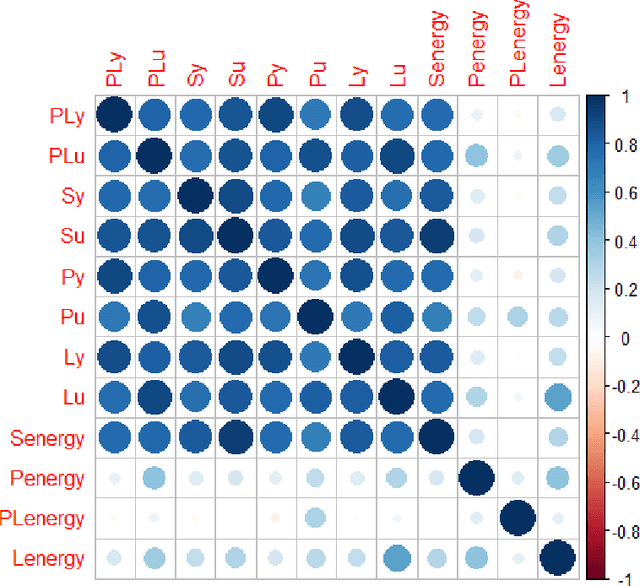
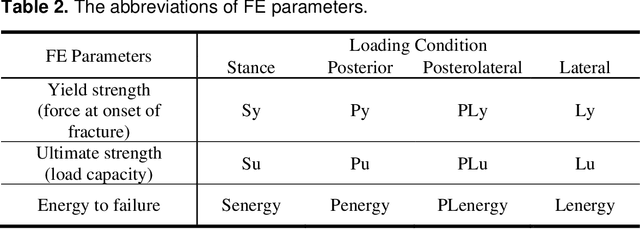
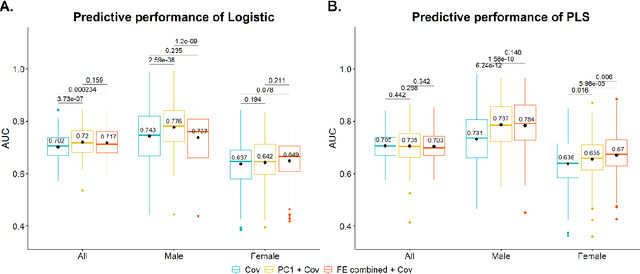
Abstract:Hip fracture risk assessment is an important but challenging task. Quantitative CT-based patient specific finite element analysis (FEA) computes the force (fracture load) to break the proximal femur in a particular loading condition. It provides different structural information about the proximal femur that can influence a subject overall fracture risk. To obtain a more robust measure of fracture risk, we used principal component analysis (PCA) to develop a global FEA computed fracture risk index that incorporates the FEA-computed yield and ultimate failure loads and energies to failure in four loading conditions (single-limb stance and impact from a fall onto the posterior, posterolateral, and lateral aspects of the greater trochanter) of 110 hip fracture subjects and 235 age and sex matched control subjects from the AGES-Reykjavik study. We found that the first PC (PC1) of the FE parameters was the only significant predictor of hip fracture. Using a logistic regression model, we determined if prediction performance for hip fracture using PC1 differed from that using FE parameters combined by stratified random resampling with respect to hip fracture status. The results showed that the average of the area under the receive operating characteristic curve (AUC) using PC1 was always higher than that using all FE parameters combined in the male subjects. The AUC of PC1 and AUC of the FE parameters combined were not significantly different than that in the female subjects or in all subjects
Multi-view information fusion using multi-view variational autoencoders to predict proximal femoral strength
Oct 03, 2022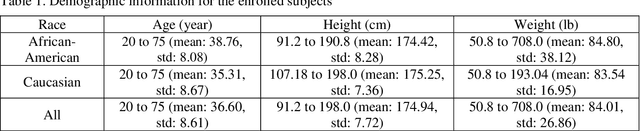
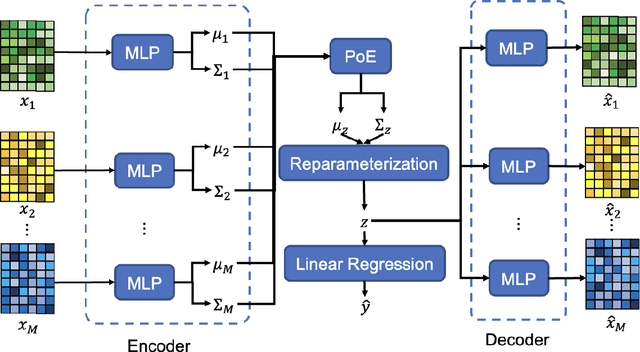
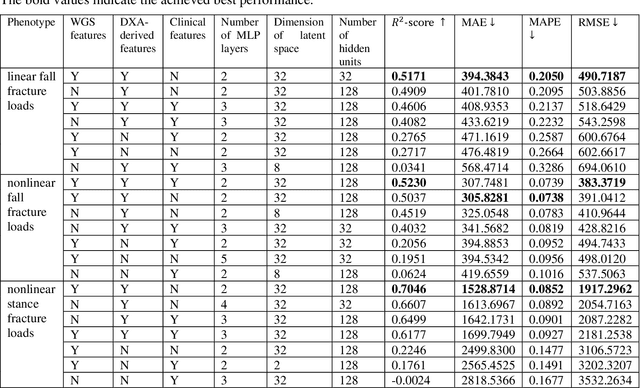
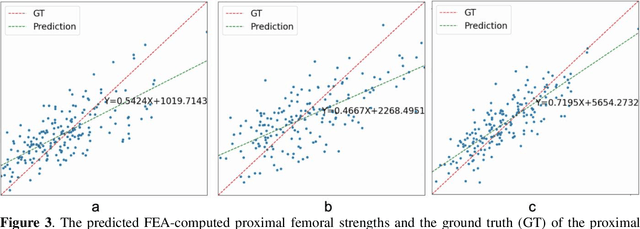
Abstract:Background and aim: Hip fracture can be devastating. The proximal femoral strength can be computed by subject-specific finite element (FE) analysis (FEA) using quantitative CT images. The aim of this paper is to design a deep learning-based model for hip fracture prediction with multi-view information fusion. Method: We developed a multi-view variational autoencoder (MMVAE) for feature representation learning and designed the product of expert model (PoE) for multi-view information fusion.We performed genome-wide association studies (GWAS) to select the most relevant genetic features with proximal femoral strengths and integrated genetic features with DXA-derived imaging features and clinical variables for proximal femoral strength prediction. Results: The designed model achieved the mean absolute percentage error of 0.2050,0.0739 and 0.0852 for linear fall, nonlinear fall and nonlinear stance fracture load prediction, respectively. For linear fall and nonlinear stance fracture load prediction, integrating genetic and DXA-derived imaging features were beneficial; while for nonlinear fall fracture load prediction, integrating genetic features, DXA-derived imaging features as well as clinical variables, the model achieved the best performance. Conclusion: The proposed model is capable of predicting proximal femoral strengths using genetic features, DXA-derived imaging features as well as clinical variables. Compared to performing FEA using QCT images to calculate proximal femoral strengths, the presented method is time-efficient and cost effective, and radiation dosage is limited. From the technique perspective, the final models can be applied to other multi-view information integration tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge