Haoyue Zhang
DA-SSL: self-supervised domain adaptor to leverage foundational models in turbt histopathology slides
Dec 15, 2025Abstract:Recent deep learning frameworks in histopathology, particularly multiple instance learning (MIL) combined with pathology foundational models (PFMs), have shown strong performance. However, PFMs exhibit limitations on certain cancer or specimen types due to domain shifts - these cancer types were rarely used for pretraining or specimens contain tissue-based artifacts rarely seen within the pretraining population. Such is the case for transurethral resection of bladder tumor (TURBT), which are essential for diagnosing muscle-invasive bladder cancer (MIBC), but contain fragmented tissue chips and electrocautery artifacts and were not widely used in publicly available PFMs. To address this, we propose a simple yet effective domain-adaptive self-supervised adaptor (DA-SSL) that realigns pretrained PFM features to the TURBT domain without fine-tuning the foundational model itself. We pilot this framework for predicting treatment response in TURBT, where histomorphological features are currently underutilized and identifying patients who will benefit from neoadjuvant chemotherapy (NAC) is challenging. In our multi-center study, DA-SSL achieved an AUC of 0.77+/-0.04 in five-fold cross-validation and an external test accuracy of 0.84, sensitivity of 0.71, and specificity of 0.91 using majority voting. Our results demonstrate that lightweight domain adaptation with self-supervision can effectively enhance PFM-based MIL pipelines for clinically challenging histopathology tasks. Code is Available at https://github.com/zhanghaoyue/DA_SSL_TURBT.
MovePose: A High-performance Human Pose Estimation Algorithm on Mobile and Edge Devices
Aug 17, 2023



Abstract:We present MovePose, an optimized lightweight convolutional neural network designed specifically for real-time body pose estimation on CPU-based mobile devices. The current solutions do not provide satisfactory accuracy and speed for human posture estimation, and MovePose addresses this gap. It aims to maintain real-time performance while improving the accuracy of human posture estimation for mobile devices. The network produces 17 keypoints for each individual at a rate exceeding 11 frames per second, making it suitable for real-time applications such as fitness tracking, sign language interpretation, and advanced mobile human posture estimation. Our MovePose algorithm has attained an Mean Average Precision (mAP) score of 67.7 on the COCO \cite{cocodata} validation dataset. The MovePose algorithm displayed efficiency with a performance of 69+ frames per second (fps) when run on an Intel i9-10920x CPU. Additionally, it showcased an increased performance of 452+ fps on an NVIDIA RTX3090 GPU. On an Android phone equipped with a Snapdragon 8 + 4G processor, the fps reached above 11. To enhance accuracy, we incorporated three techniques: deconvolution, large kernel convolution, and coordinate classification methods. Compared to basic upsampling, deconvolution is trainable, improves model capacity, and enhances the receptive field. Large kernel convolution strengthens these properties at a decreased computational cost. In summary, MovePose provides high accuracy and real-time performance, marking it a potential tool for a variety of applications, including those focused on mobile-side human posture estimation. The code and models for this algorithm will be made publicly accessible.
Predicting Thrombectomy Recanalization from CT Imaging Using Deep Learning Models
Feb 08, 2023Abstract:For acute ischemic stroke (AIS) patients with large vessel occlusions, clinicians must decide if the benefit of mechanical thrombectomy (MTB) outweighs the risks and potential complications following an invasive procedure. Pre-treatment computed tomography (CT) and angiography (CTA) are widely used to characterize occlusions in the brain vasculature. If a patient is deemed eligible, a modified treatment in cerebral ischemia (mTICI) score will be used to grade how well blood flow is reestablished throughout and following the MTB procedure. An estimation of the likelihood of successful recanalization can support treatment decision-making. In this study, we proposed a fully automated prediction of a patient's recanalization score using pre-treatment CT and CTA imaging. We designed a spatial cross attention network (SCANet) that utilizes vision transformers to localize to pertinent slices and brain regions. Our top model achieved an average cross-validated ROC-AUC of 77.33 $\pm$ 3.9\%. This is a promising result that supports future applications of deep learning on CT and CTA for the identification of eligible AIS patients for MTB.
* Medical Imaging with Deep Learning 2022 accepted short paper Jun 2022
MMMNA-Net for Overall Survival Time Prediction of Brain Tumor Patients
Jun 13, 2022



Abstract:Overall survival (OS) time is one of the most important evaluation indices for gliomas situations. Multimodal Magnetic Resonance Imaging (MRI) scans play an important role in the study of glioma prognosis OS time. Several deep learning-based methods are proposed for the OS time prediction on multi-modal MRI problems. However, these methods usually fuse multi-modal information at the beginning or at the end of the deep learning networks and lack the fusion of features from different scales. In addition, the fusion at the end of networks always adapts global with global (eg. fully connected after concatenation of global average pooling output) or local with local (eg. bilinear pooling), which loses the information of local with global. In this paper, we propose a novel method for multi-modal OS time prediction of brain tumor patients, which contains an improved nonlocal features fusion module introduced on different scales. Our method obtains a relative 8.76% improvement over the current state-of-art method (0.6989 vs. 0.6426 on accuracy). Extensive testing demonstrates that our method could adapt to situations with missing modalities. The code is available at https://github.com/TangWen920812/mmmna-net.
RPLHR-CT Dataset and Transformer Baseline for Volumetric Super-Resolution from CT Scans
Jun 13, 2022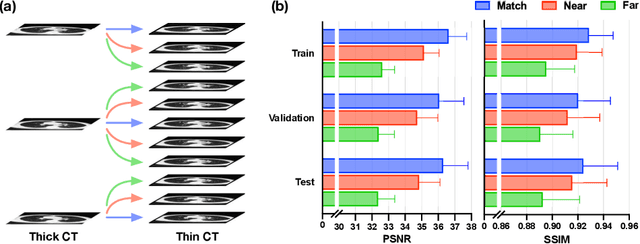
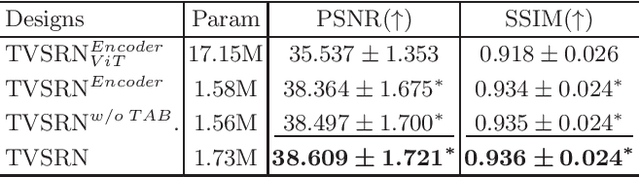
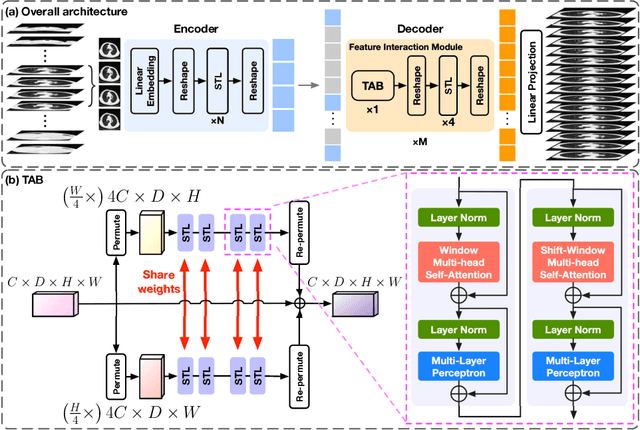
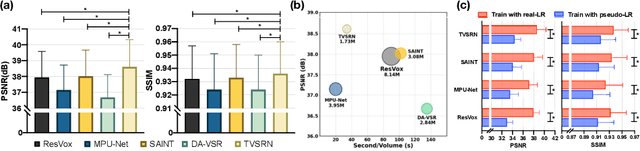
Abstract:In clinical practice, anisotropic volumetric medical images with low through-plane resolution are commonly used due to short acquisition time and lower storage cost. Nevertheless, the coarse resolution may lead to difficulties in medical diagnosis by either physicians or computer-aided diagnosis algorithms. Deep learning-based volumetric super-resolution (SR) methods are feasible ways to improve resolution, with convolutional neural networks (CNN) at their core. Despite recent progress, these methods are limited by inherent properties of convolution operators, which ignore content relevance and cannot effectively model long-range dependencies. In addition, most of the existing methods use pseudo-paired volumes for training and evaluation, where pseudo low-resolution (LR) volumes are generated by a simple degradation of their high-resolution (HR) counterparts. However, the domain gap between pseudo- and real-LR volumes leads to the poor performance of these methods in practice. In this paper, we build the first public real-paired dataset RPLHR-CT as a benchmark for volumetric SR, and provide baseline results by re-implementing four state-of-the-art CNN-based methods. Considering the inherent shortcoming of CNN, we also propose a transformer volumetric super-resolution network (TVSRN) based on attention mechanisms, dispensing with convolutions entirely. This is the first research to use a pure transformer for CT volumetric SR. The experimental results show that TVSRN significantly outperforms all baselines on both PSNR and SSIM. Moreover, the TVSRN method achieves a better trade-off between the image quality, the number of parameters, and the running time. Data and code are available at https://github.com/smilenaxx/RPLHR-CT.
Transformer Lesion Tracker
Jun 13, 2022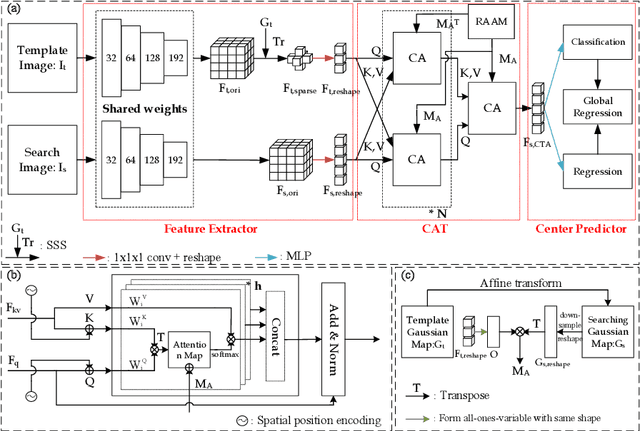
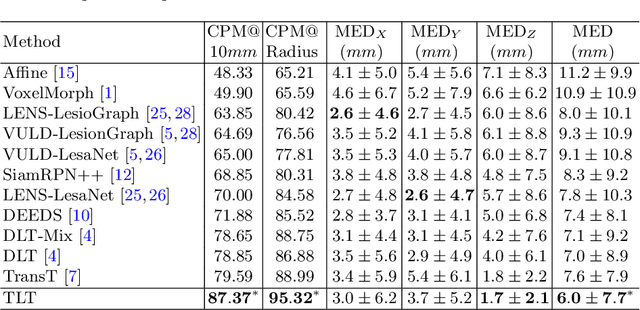
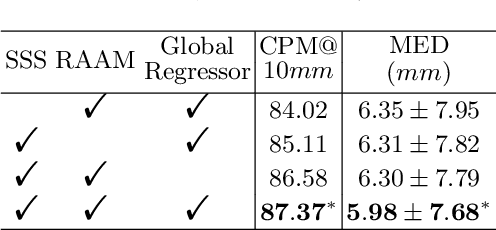
Abstract:Evaluating lesion progression and treatment response via longitudinal lesion tracking plays a critical role in clinical practice. Automated approaches for this task are motivated by prohibitive labor costs and time consumption when lesion matching is done manually. Previous methods typically lack the integration of local and global information. In this work, we propose a transformer-based approach, termed Transformer Lesion Tracker (TLT). Specifically, we design a Cross Attention-based Transformer (CAT) to capture and combine both global and local information to enhance feature extraction. We also develop a Registration-based Anatomical Attention Module (RAAM) to introduce anatomical information to CAT so that it can focus on useful feature knowledge. A Sparse Selection Strategy (SSS) is presented for selecting features and reducing memory footprint in Transformer training. In addition, we use a global regression to further improve model performance. We conduct experiments on a public dataset to show the superiority of our method and find that our model performance has improved the average Euclidean center error by at least 14.3% (6mm vs. 7mm) compared with the state-of-the-art (SOTA). Code is available at https://github.com/TangWen920812/TLT.
Intra-Domain Task-Adaptive Transfer Learning to Determine Acute Ischemic Stroke Onset Time
Nov 05, 2020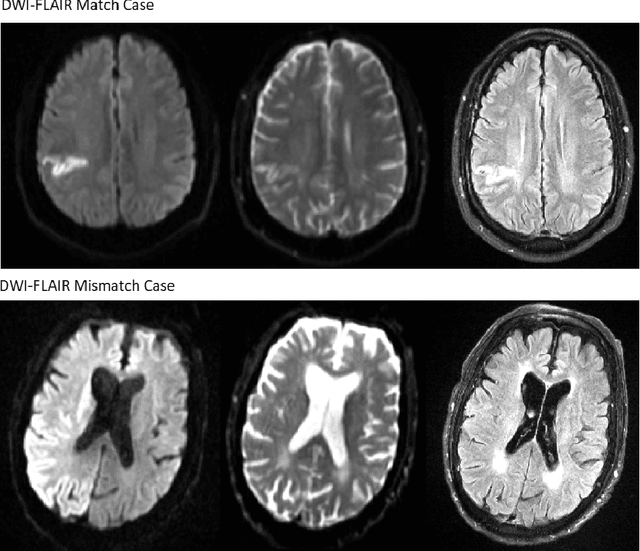
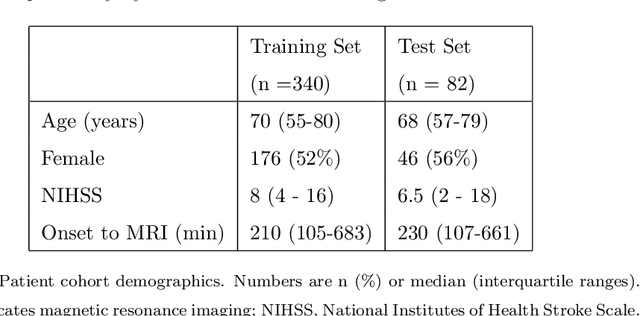
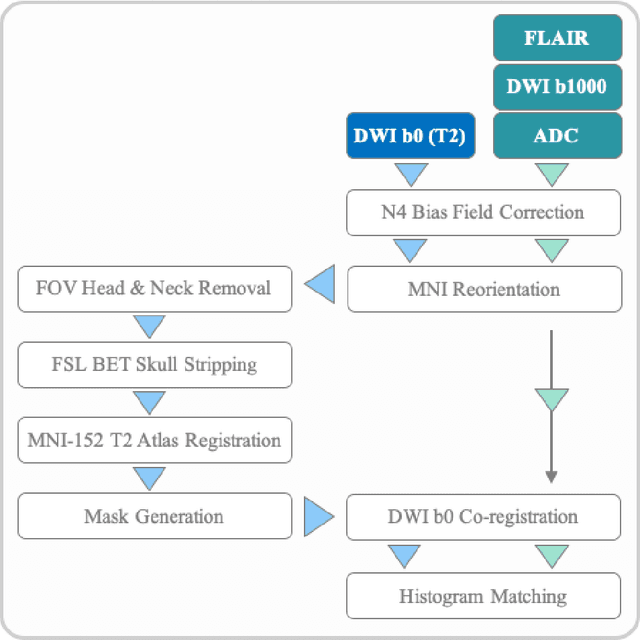
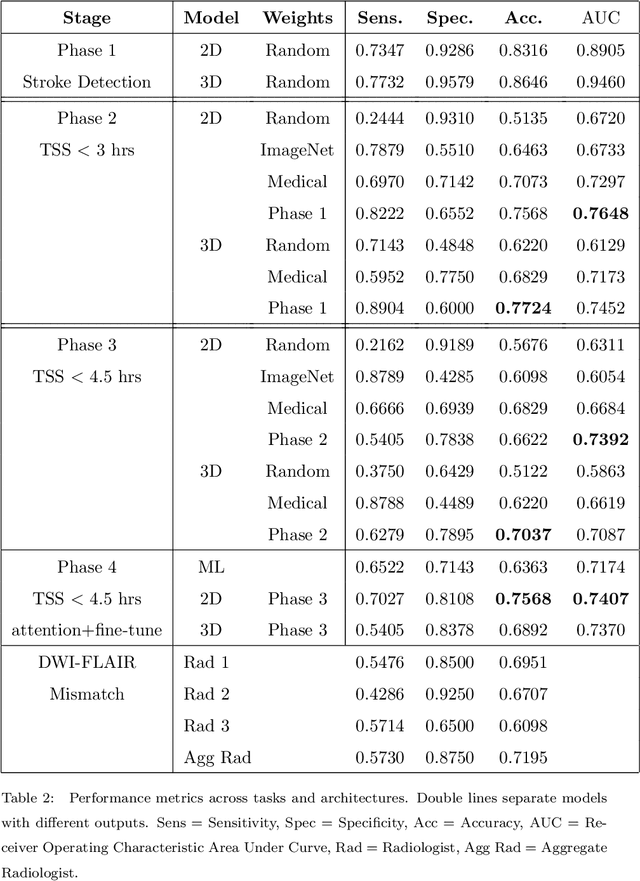
Abstract:Treatment of acute ischemic strokes (AIS) is largely contingent upon the time since stroke onset (TSS). However, TSS may not be readily available in up to 25% of patients with unwitnessed AIS. Current clinical guidelines for patients with unknown TSS recommend the use of MRI to determine eligibility for thrombolysis, but radiology assessments have high inter-reader variability. In this work, we present deep learning models that leverage MRI diffusion series to classify TSS based on clinically validated thresholds. We propose an intra-domain task-adaptive transfer learning method, which involves training a model on an easier clinical task (stroke detection) and then refining the model with different binary thresholds of TSS. We apply this approach to both 2D and 3D CNN architectures with our top model achieving an ROC-AUC value of 0.74, with a sensitivity of 0.70 and a specificity of 0.81 for classifying TSS < 4.5 hours. Our pretrained models achieve better classification metrics than the models trained from scratch, and these metrics exceed those of previously published models applied to our dataset. Furthermore, our pipeline accommodates a more inclusive patient cohort than previous work, as we did not exclude imaging studies based on clinical, demographic, or image processing criteria. When applied to this broad spectrum of patients, our deep learning model achieves an overall accuracy of 75.78% when classifying TSS < 4.5 hours, carrying potential therapeutic implications for patients with unknown TSS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge