Ashwath Radhachandran
CytoFM: The first cytology foundation model
Apr 18, 2025Abstract:Cytology is essential for cancer diagnostics and screening due to its minimally invasive nature. However, the development of robust deep learning models for digital cytology is challenging due to the heterogeneity in staining and preparation methods of samples, differences across organs, and the limited availability of large, diverse, annotated datasets. Developing a task-specific model for every cytology application is impractical and non-cytology-specific foundation models struggle to generalize to tasks in this domain where the emphasis is on cell morphology. To address these challenges, we introduce CytoFM, the first cytology self-supervised foundation model. Using iBOT, a self-supervised Vision Transformer (ViT) training framework incorporating masked image modeling and self-distillation, we pretrain CytoFM on a diverse collection of cytology datasets to learn robust, transferable representations. We evaluate CytoFM on multiple downstream cytology tasks, including breast cancer classification and cell type identification, using an attention-based multiple instance learning framework. Our results demonstrate that CytoFM performs better on two out of three downstream tasks than existing foundation models pretrained on histopathology (UNI) or natural images (iBOT-Imagenet). Visualizations of learned representations demonstrate our model is able to attend to cytologically relevant features. Despite a small pre-training dataset, CytoFM's promising results highlight the ability of task-agnostic pre-training approaches to learn robust and generalizable features from cytology data.
Reducing Overtreatment of Indeterminate Thyroid Nodules Using a Multimodal Deep Learning Model
Sep 27, 2024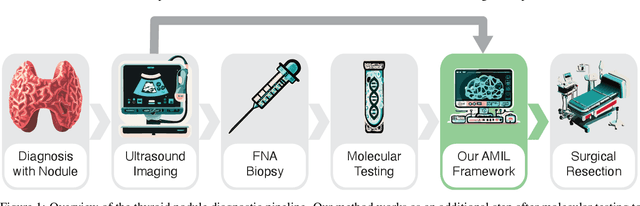
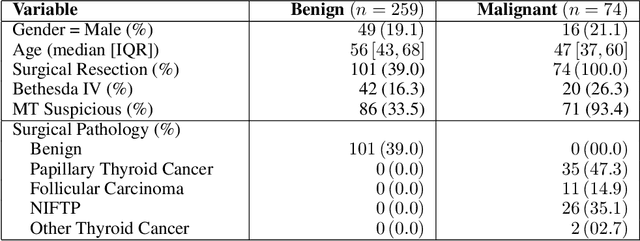
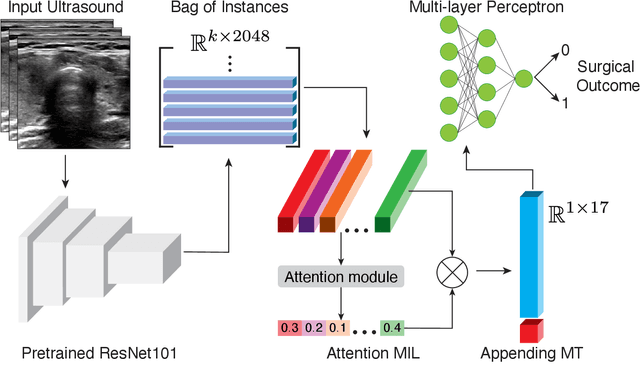
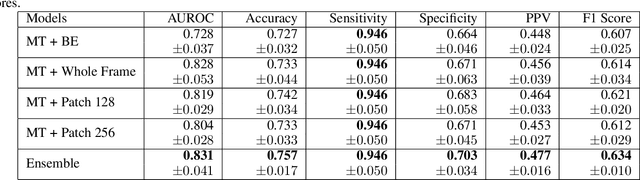
Abstract:Objective: Molecular testing (MT) classifies cytologically indeterminate thyroid nodules as benign or malignant with high sensitivity but low positive predictive value (PPV), only using molecular profiles, ignoring ultrasound (US) imaging and biopsy. We address this limitation by applying attention multiple instance learning (AMIL) to US images. Methods: We retrospectively reviewed 333 patients with indeterminate thyroid nodules at UCLA medical center (259 benign, 74 malignant). A multi-modal deep learning AMIL model was developed, combining US images and MT to classify the nodules as benign or malignant and enhance the malignancy risk stratification of MT. Results: The final AMIL model matched MT sensitivity (0.946) while significantly improving PPV (0.477 vs 0.448 for MT alone), indicating fewer false positives while maintaining high sensitivity. Conclusion: Our approach reduces false positives compared to MT while maintaining the same ability to identify positive cases, potentially reducing unnecessary benign thyroid resections in patients with indeterminate nodules.
Ultrasound Image Enhancement using CycleGAN and Perceptual Loss
Dec 18, 2023Abstract:Purpose: The objective of this work is to introduce an advanced framework designed to enhance ultrasound images, especially those captured by portable hand-held devices, which often produce lower quality images due to hardware constraints. Additionally, this framework is uniquely capable of effectively handling non-registered input ultrasound image pairs, addressing a common challenge in medical imaging. Materials and Methods: In this retrospective study, we utilized an enhanced generative adversarial network (CycleGAN) model for ultrasound image enhancement across five organ systems. Perceptual loss, derived from deep features of pretrained neural networks, is applied to ensure the human-perceptual quality of the enhanced images. These images are compared with paired images acquired from high resolution devices to demonstrate the model's ability to generate realistic high-quality images across organ systems. Results: Preliminary validation of the framework reveals promising performance metrics. The model generates images that result in a Structural Similarity Index (SSI) score of 0.722, Locally Normalized Cross-Correlation (LNCC) score of 0.902 and 28.802 for the Peak Signal-to-Noise Ratio (PSNR) metric. Conclusion: This work presents a significant advancement in medical imaging through the development of a CycleGAN model enhanced with Perceptual Loss (PL), effectively bridging the quality gap between ultrasound images from varied devices. By training on paired images, the model not only improves image quality but also ensures the preservation of vital anatomic structural content. This approach may improve equity in access to healthcare by enhancing portable device capabilities, although further validation and optimizations are necessary for broader clinical application.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge