Abhejit Rajagopal
Mixed Supervision of Histopathology Improves Prostate Cancer Classification from MRI
Dec 13, 2022



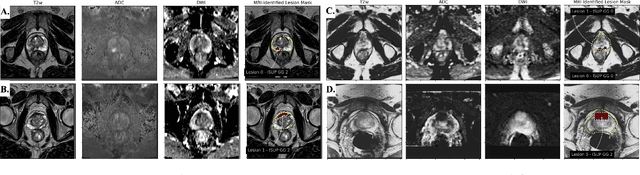
Abstract:Non-invasive prostate cancer detection from MRI has the potential to revolutionize patient care by providing early detection of clinically-significant disease (ISUP grade group >= 2), but has thus far shown limited positive predictive value. To address this, we present an MRI-based deep learning method for predicting clinically significant prostate cancer applicable to a patient population with subsequent ground truth biopsy results ranging from benign pathology to ISUP grade group~5. Specifically, we demonstrate that mixed supervision via diverse histopathological ground truth improves classification performance despite the cost of reduced concordance with image-based segmentation. That is, where prior approaches have utilized pathology results as ground truth derived from targeted biopsies and whole-mount prostatectomy to strongly supervise the localization of clinically significant cancer, our approach also utilizes weak supervision signals extracted from nontargeted systematic biopsies with regional localization to improve overall performance. Our key innovation is performing regression by distribution rather than simply by value, enabling use of additional pathology findings traditionally ignored by deep learning strategies. We evaluated our model on a dataset of 973 (testing n=160) multi-parametric prostate MRI exams collected at UCSF from 2015-2018 followed by MRI/ultrasound fusion (targeted) biopsy and systematic (nontargeted) biopsy of the prostate gland, demonstrating that deep networks trained with mixed supervision of histopathology can significantly exceed the performance of the Prostate Imaging-Reporting and Data System (PI-RADS) clinical standard for prostate MRI interpretation.
Physics-driven Deep Learning for PET/MRI
Jun 11, 2022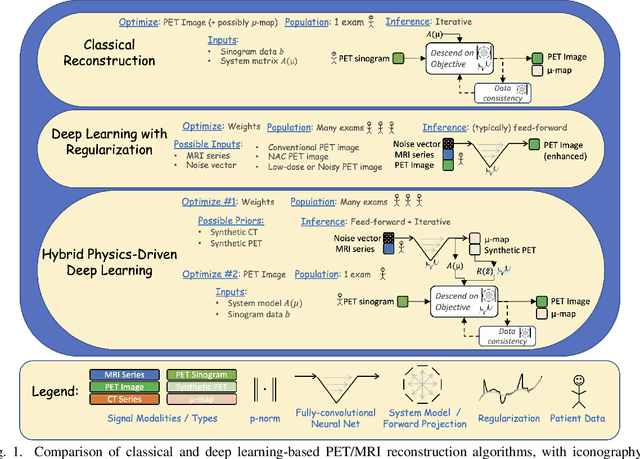
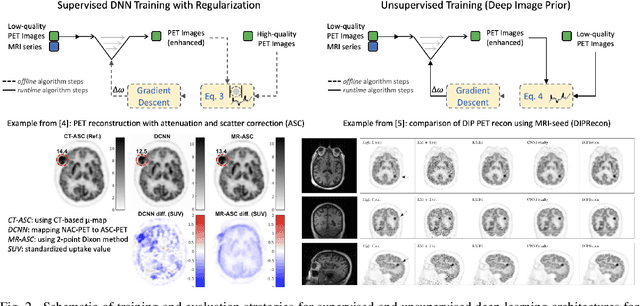
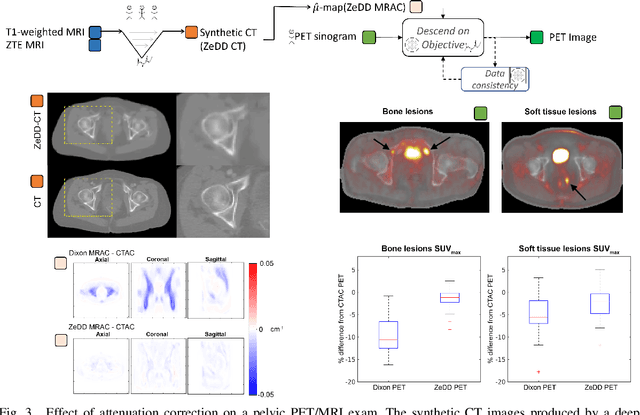
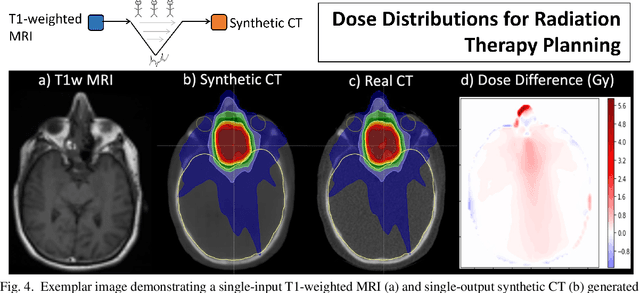
Abstract:In this paper, we review physics- and data-driven reconstruction techniques for simultaneous positron emission tomography (PET) / magnetic resonance imaging (MRI) systems, which have significant advantages for clinical imaging of cancer, neurological disorders, and heart disease. These reconstruction approaches utilize priors, either structural or statistical, together with a physics-based description of the PET system response. However, due to the nested representation of the forward problem, direct PET/MRI reconstruction is a nonlinear problem. We elucidate how a multi-faceted approach accommodates hybrid data- and physics-driven machine learning for reconstruction of 3D PET/MRI, summarizing important deep learning developments made in the last 5 years to address attenuation correction, scattering, low photon counts, and data consistency. We also describe how applications of these multi-modality approaches extend beyond PET/MRI to improving accuracy in radiation therapy planning. We conclude by discussing opportunities for extending the current state-of-the-art following the latest trends in physics- and deep learning-based computational imaging and next-generation detector hardware.
Synthetic PET via Domain Translation of 3D MRI
Jun 11, 2022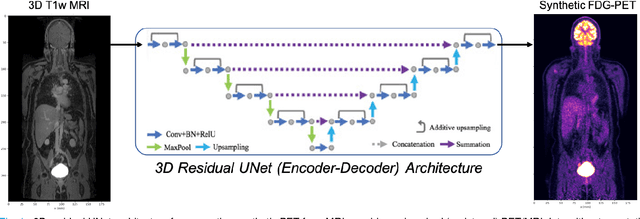
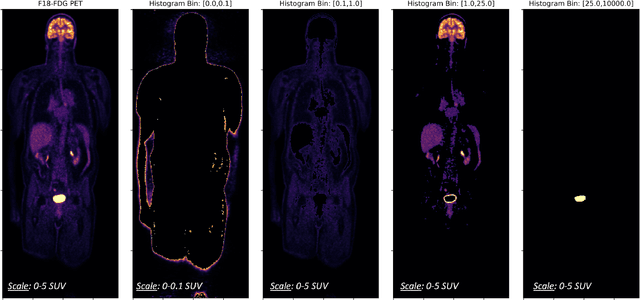
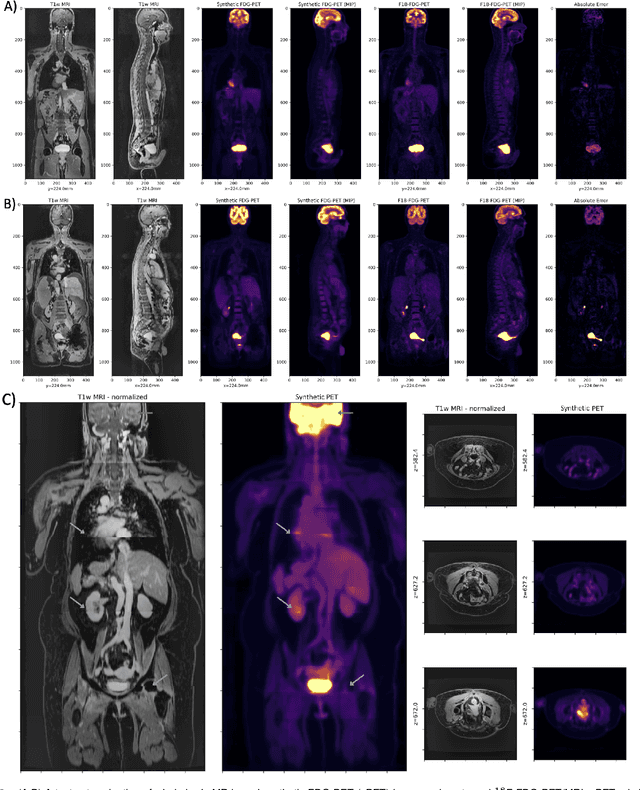
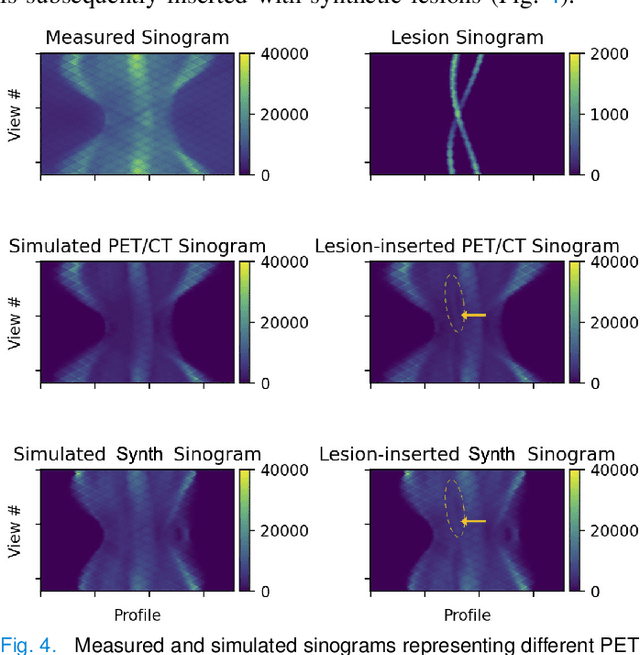
Abstract:Historically, patient datasets have been used to develop and validate various reconstruction algorithms for PET/MRI and PET/CT. To enable such algorithm development, without the need for acquiring hundreds of patient exams, in this paper we demonstrate a deep learning technique to generate synthetic but realistic whole-body PET sinograms from abundantly-available whole-body MRI. Specifically, we use a dataset of 56 $^{18}$F-FDG-PET/MRI exams to train a 3D residual UNet to predict physiologic PET uptake from whole-body T1-weighted MRI. In training we implemented a balanced loss function to generate realistic uptake across a large dynamic range and computed losses along tomographic lines of response to mimic the PET acquisition. The predicted PET images are forward projected to produce synthetic PET time-of-flight (ToF) sinograms that can be used with vendor-provided PET reconstruction algorithms, including using CT-based attenuation correction (CTAC) and MR-based attenuation correction (MRAC). The resulting synthetic data recapitulates physiologic $^{18}$F-FDG uptake, e.g. high uptake localized to the brain and bladder, as well as uptake in liver, kidneys, heart and muscle. To simulate abnormalities with high uptake, we also insert synthetic lesions. We demonstrate that this synthetic PET data can be used interchangeably with real PET data for the PET quantification task of comparing CT and MR-based attenuation correction methods, achieving $\leq 7.6\%$ error in mean-SUV compared to using real data. These results together show that the proposed synthetic PET data pipeline can be reasonably used for development, evaluation, and validation of PET/MRI reconstruction methods.
Federated Learning with Research Prototypes for Multi-Center MRI-based Detection of Prostate Cancer with Diverse Histopathology
Jun 11, 2022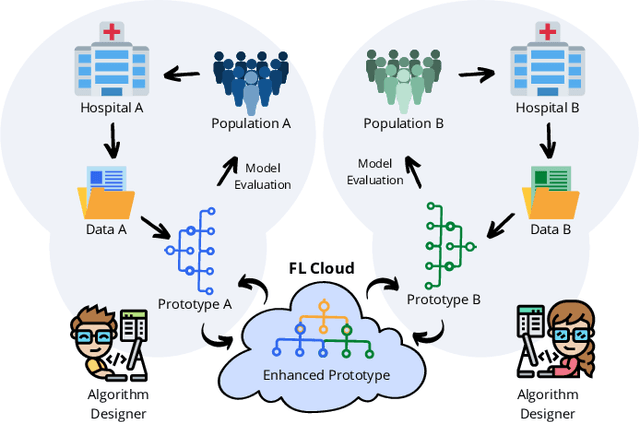
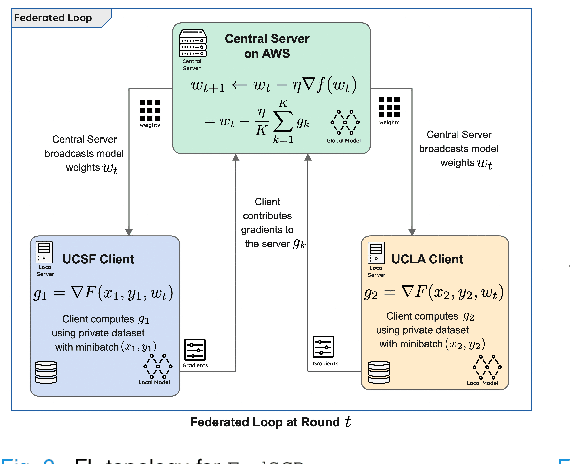
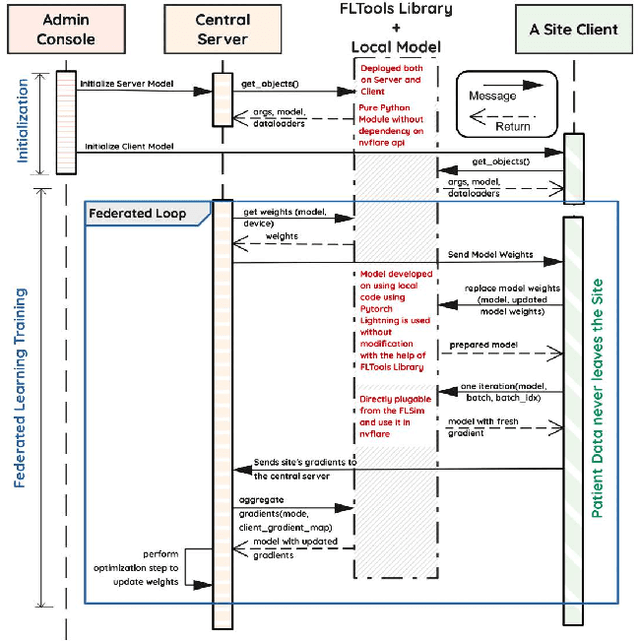
Abstract:Early prostate cancer detection and staging from MRI are extremely challenging tasks for both radiologists and deep learning algorithms, but the potential to learn from large and diverse datasets remains a promising avenue to increase their generalization capability both within- and across clinics. To enable this for prototype-stage algorithms, where the majority of existing research remains, in this paper we introduce a flexible federated learning framework for cross-site training, validation, and evaluation of deep prostate cancer detection algorithms. Our approach utilizes an abstracted representation of the model architecture and data, which allows unpolished prototype deep learning models to be trained without modification using the NVFlare federated learning framework. Our results show increases in prostate cancer detection and classification accuracy using a specialized neural network model and diverse prostate biopsy data collected at two University of California research hospitals, demonstrating the efficacy of our approach in adapting to different datasets and improving MR-biomarker discovery. We open-source our FLtools system, which can be easily adapted to other deep learning projects for medical imaging.
Predicting Generalization in Deep Learning via Local Measures of Distortion
Dec 16, 2020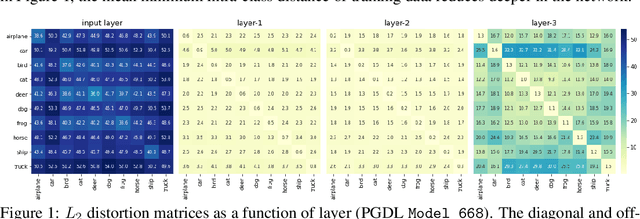
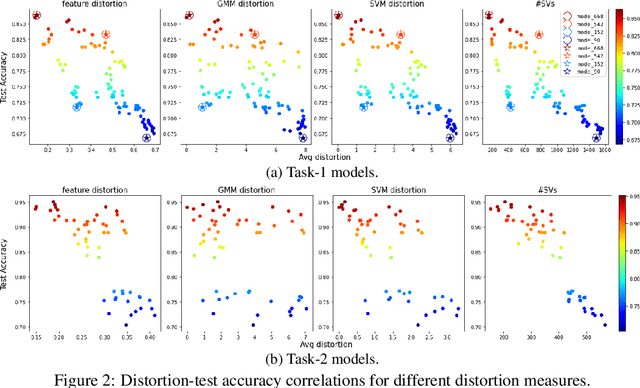
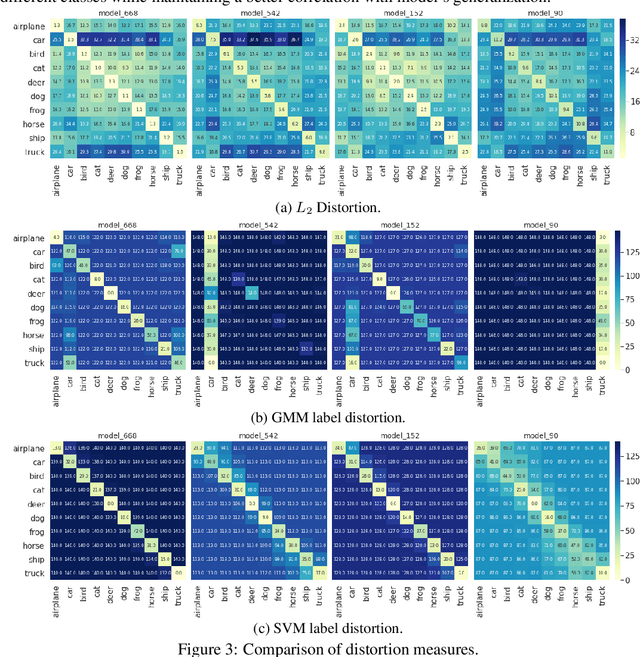
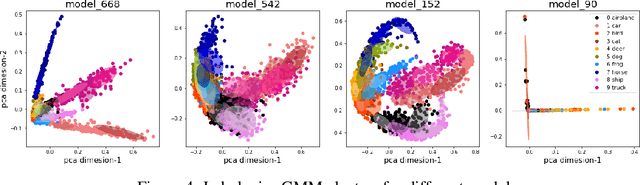
Abstract:We study generalization in deep learning by appealing to complexity measures originally developed in approximation and information theory. While these concepts are challenged by the high-dimensional and data-defined nature of deep learning, we show that simple vector quantization approaches such as PCA, GMMs, and SVMs capture their spirit when applied layer-wise to deep extracted features giving rise to relatively inexpensive complexity measures that correlate well with generalization performance. We discuss our results in 2020 NeurIPS PGDL challenge.
Deep Algorithms: designs for networks
Jun 06, 2018



Abstract:A new design methodology for neural networks that is guided by traditional algorithm design is presented. To prove our point, we present two heuristics and demonstrate an algorithmic technique for incorporating additional weights in their signal-flow graphs. We show that with training the performance of these networks can not only exceed the performance of the initial network, but can match the performance of more-traditional neural network architectures. A key feature of our approach is that these networks are initialized with parameters that provide a known performance threshold for the architecture on a given task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge