Richard Laforest
From Diagnosis to Therapy: Progress in SPECT and PET Reconstruction for Theranostics
Sep 09, 2025Abstract:The theranostic paradigm enables personalization of treatment by selecting patients with a diagnostic radiopharmaceutical and monitoring therapy using a matched therapeutic isotope. This strategy relies on accurate image reconstruction of both pre-therapy and post-therapy images for patient selection and monitoring treatment. However, traditional reconstruction methods are hindered by challenges such as crosstalk in multi-isotope imaging and extremely low-count measurements when imaging of alpha- ({\alpha}-) emitting therapies. Additionally, to fully realize the benefits of new imaging systems being developed for theranostic applications, advanced reconstruction techniques are needed. These needs, alongside the growing clinical adoption of theranostics, have spurred the development of novel PET and SPECT reconstruction algorithms. This review highlights recent progress and addresses critical challenges and unmet needs in theranostic image reconstruction.
DEMIST: A deep-learning-based task-specific denoising approach for myocardial perfusion SPECT
Jun 14, 2023Abstract:There is an important need for methods to process myocardial perfusion imaging (MPI) SPECT images acquired at lower radiation dose and/or acquisition time such that the processed images improve observer performance on the clinical task of detecting perfusion defects. To address this need, we build upon concepts from model-observer theory and our understanding of the human visual system to propose a Detection task-specific deep-learning-based approach for denoising MPI SPECT images (DEMIST). The approach, while performing denoising, is designed to preserve features that influence observer performance on detection tasks. We objectively evaluated DEMIST on the task of detecting perfusion defects using a retrospective study with anonymized clinical data in patients who underwent MPI studies across two scanners (N = 338). The evaluation was performed at low-dose levels of 6.25%, 12.5% and 25% and using an anthropomorphic channelized Hotelling observer. Performance was quantified using area under the receiver operating characteristics curve (AUC). Images denoised with DEMIST yielded significantly higher AUC compared to corresponding low-dose images and images denoised with a commonly used task-agnostic DL-based denoising method. Similar results were observed with stratified analysis based on patient sex and defect type. Additionally, DEMIST improved visual fidelity of the low-dose images as quantified using root mean squared error and structural similarity index metric. A mathematical analysis revealed that DEMIST preserved features that assist in detection tasks while improving the noise properties, resulting in improved observer performance. The results provide strong evidence for further clinical evaluation of DEMIST to denoise low-count images in MPI SPECT.
Need for Objective Task-based Evaluation of Deep Learning-Based Denoising Methods: A Study in the Context of Myocardial Perfusion SPECT
Mar 16, 2023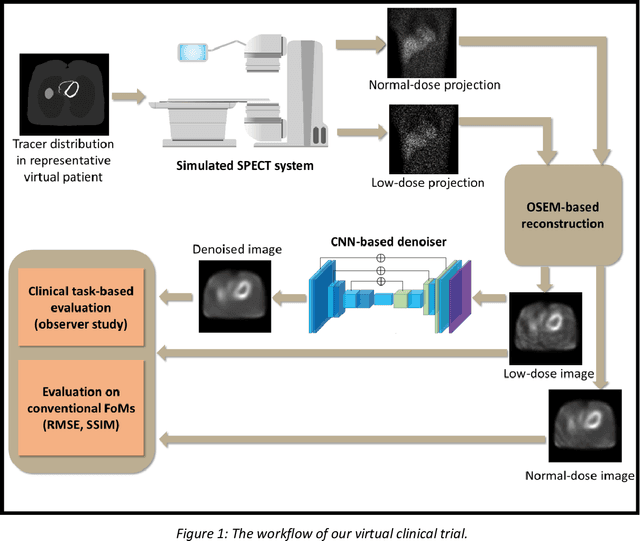
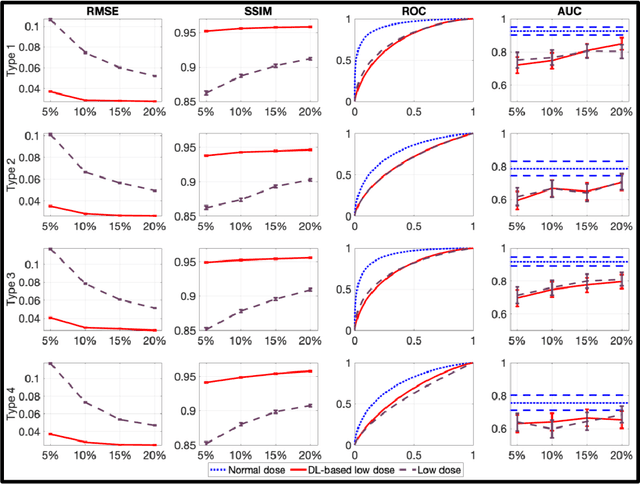
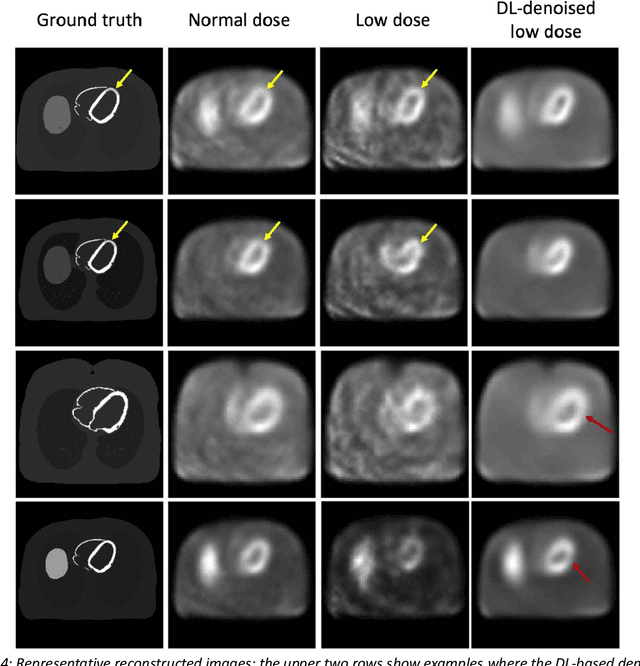
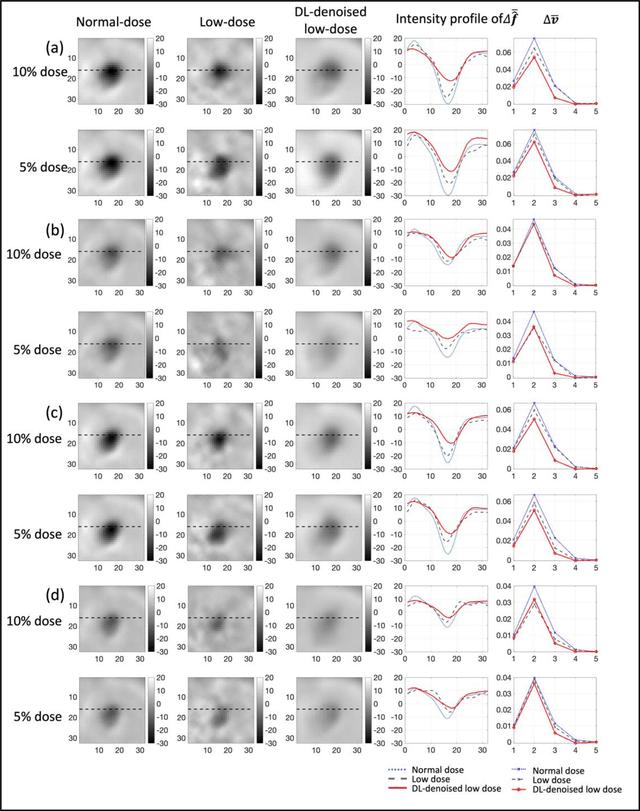
Abstract:Artificial intelligence-based methods have generated substantial interest in nuclear medicine. An area of significant interest has been using deep-learning (DL)-based approaches for denoising images acquired with lower doses, shorter acquisition times, or both. Objective evaluation of these approaches is essential for clinical application. DL-based approaches for denoising nuclear-medicine images have typically been evaluated using fidelity-based figures of merit (FoMs) such as RMSE and SSIM. However, these images are acquired for clinical tasks and thus should be evaluated based on their performance in these tasks. Our objectives were to (1) investigate whether evaluation with these FoMs is consistent with objective clinical-task-based evaluation; (2) provide a theoretical analysis for determining the impact of denoising on signal-detection tasks; (3) demonstrate the utility of virtual clinical trials (VCTs) to evaluate DL-based methods. A VCT to evaluate a DL-based method for denoising myocardial perfusion SPECT (MPS) images was conducted. The impact of DL-based denoising was evaluated using fidelity-based FoMs and AUC, which quantified performance on detecting perfusion defects in MPS images as obtained using a model observer with anthropomorphic channels. Based on fidelity-based FoMs, denoising using the considered DL-based method led to significantly superior performance. However, based on ROC analysis, denoising did not improve, and in fact, often degraded detection-task performance. The results motivate the need for objective task-based evaluation of DL-based denoising approaches. Further, this study shows how VCTs provide a mechanism to conduct such evaluations using VCTs. Finally, our theoretical treatment reveals insights into the reasons for the limited performance of the denoising approach.
Synthetic PET via Domain Translation of 3D MRI
Jun 11, 2022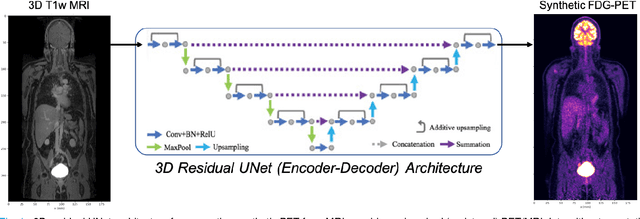
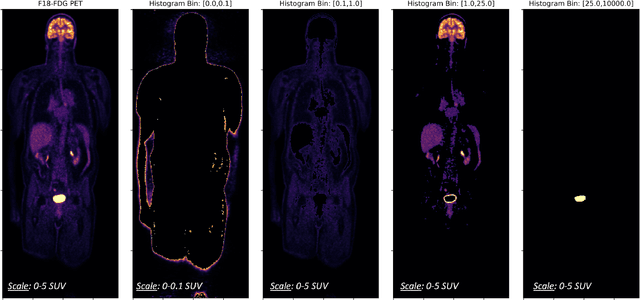
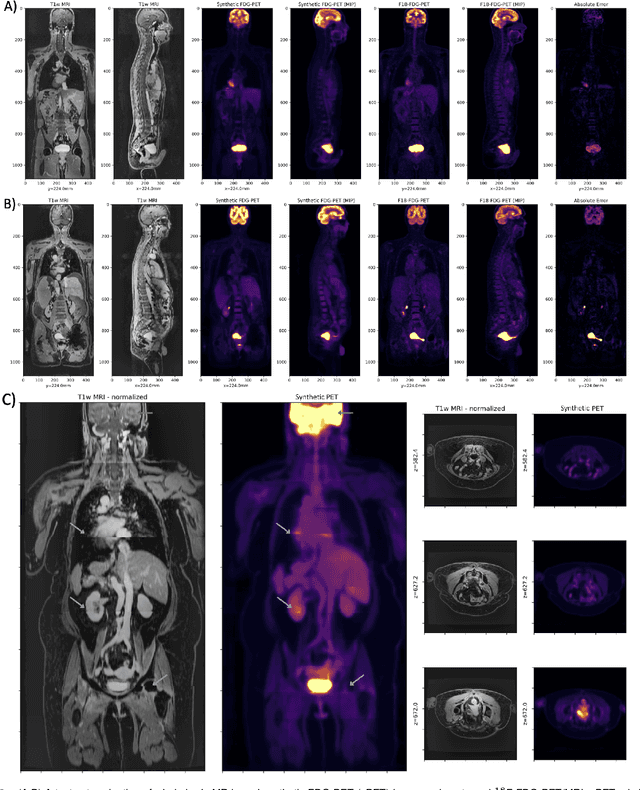
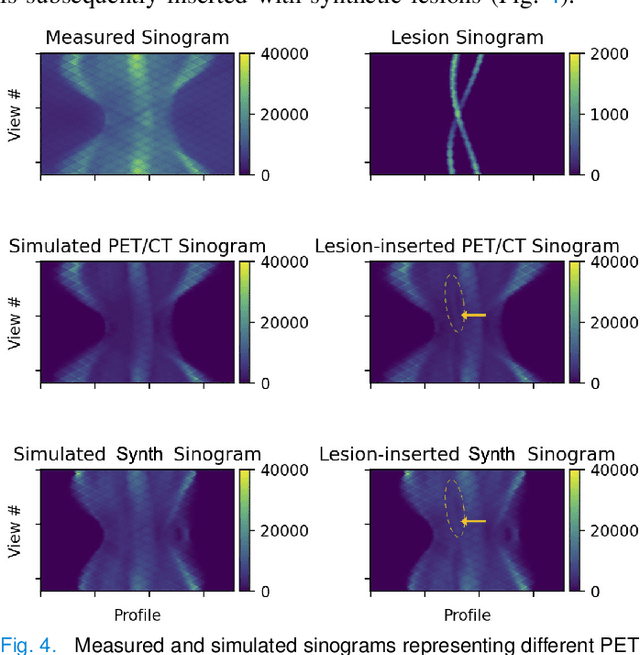
Abstract:Historically, patient datasets have been used to develop and validate various reconstruction algorithms for PET/MRI and PET/CT. To enable such algorithm development, without the need for acquiring hundreds of patient exams, in this paper we demonstrate a deep learning technique to generate synthetic but realistic whole-body PET sinograms from abundantly-available whole-body MRI. Specifically, we use a dataset of 56 $^{18}$F-FDG-PET/MRI exams to train a 3D residual UNet to predict physiologic PET uptake from whole-body T1-weighted MRI. In training we implemented a balanced loss function to generate realistic uptake across a large dynamic range and computed losses along tomographic lines of response to mimic the PET acquisition. The predicted PET images are forward projected to produce synthetic PET time-of-flight (ToF) sinograms that can be used with vendor-provided PET reconstruction algorithms, including using CT-based attenuation correction (CTAC) and MR-based attenuation correction (MRAC). The resulting synthetic data recapitulates physiologic $^{18}$F-FDG uptake, e.g. high uptake localized to the brain and bladder, as well as uptake in liver, kidneys, heart and muscle. To simulate abnormalities with high uptake, we also insert synthetic lesions. We demonstrate that this synthetic PET data can be used interchangeably with real PET data for the PET quantification task of comparing CT and MR-based attenuation correction methods, achieving $\leq 7.6\%$ error in mean-SUV compared to using real data. These results together show that the proposed synthetic PET data pipeline can be reasonably used for development, evaluation, and validation of PET/MRI reconstruction methods.
Observer study-based evaluation of a stochastic and physics-based method to generate oncological PET images
Feb 11, 2021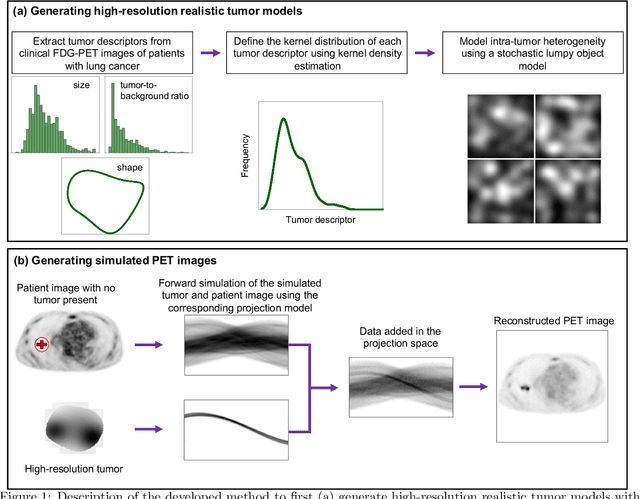


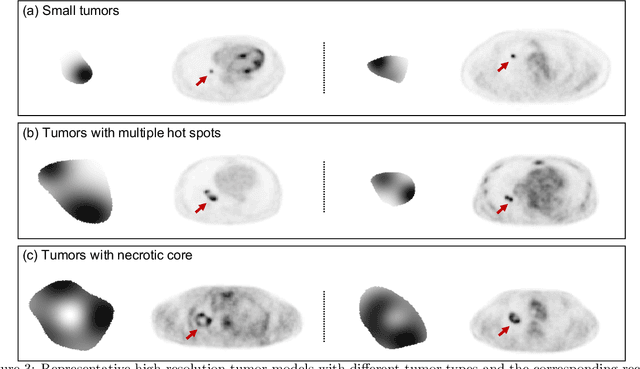
Abstract:Objective evaluation of new and improved methods for PET imaging requires access to images with ground truth, as can be obtained through simulation studies. However, for these studies to be clinically relevant, it is important that the simulated images are clinically realistic. In this study, we develop a stochastic and physics-based method to generate realistic oncological two-dimensional (2-D) PET images, where the ground-truth tumor properties are known. The developed method extends upon a previously proposed approach. The approach captures the observed variabilities in tumor properties from actual patient population. Further, we extend that approach to model intra-tumor heterogeneity using a lumpy object model. To quantitatively evaluate the clinical realism of the simulated images, we conducted a human-observer study. This was a two-alternative forced-choice (2AFC) study with trained readers (five PET physicians and one PET physicist). Our results showed that the readers had an average of ~ 50% accuracy in the 2AFC study. Further, the developed simulation method was able to generate wide varieties of clinically observed tumor types. These results provide evidence for the application of this method to 2-D PET imaging applications, and motivate development of this method to generate 3-D PET images.
Fully automated 3D segmentation of dopamine transporter SPECT images using an estimation-based approach
Jan 17, 2021



Abstract:Quantitative measures of uptake in caudate, putamen, and globus pallidus in dopamine transporter (DaT) brain SPECT have potential as biomarkers for the severity of Parkinson disease. Reliable quantification of uptake requires accurate segmentation of these regions. However, segmentation is challenging in DaT SPECT due to partial-volume effects, system noise, physiological variability, and the small size of these regions. To address these challenges, we propose an estimation-based approach to segmentation. This approach estimates the posterior mean of the fractional volume occupied by caudate, putamen, and globus pallidus within each voxel of a 3D SPECT image. The estimate is obtained by minimizing a cost function based on the binary cross-entropy loss between the true and estimated fractional volumes over a population of SPECT images, where the distribution of the true fractional volumes is obtained from magnetic resonance images from clinical populations. The proposed method accounts for both the sources of partial-volume effects in SPECT, namely the limited system resolution and tissue-fraction effects. The method was implemented using an encoder-decoder network and evaluated using realistic clinically guided SPECT simulation studies, where the ground-truth fractional volumes were known. The method significantly outperformed all other considered segmentation methods and yielded accurate segmentation with dice similarity coefficients of ~ 0.80 for all regions. The method was relatively insensitive to changes in voxel size. Further, the method was relatively robust up to +/- 10 degrees of patient head tilt along transaxial, sagittal, and coronal planes. Overall, the results demonstrate the efficacy of the proposed method to yield accurate fully automated segmentation of caudate, putamen, and globus pallidus in 3D DaT-SPECT images.
An estimation-based method to segment PET images
Feb 29, 2020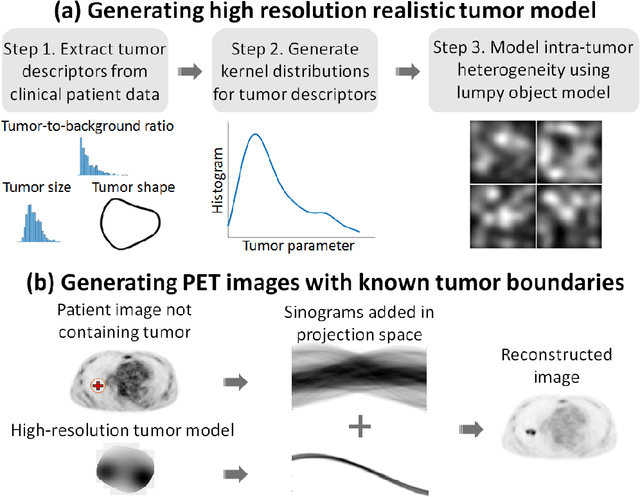
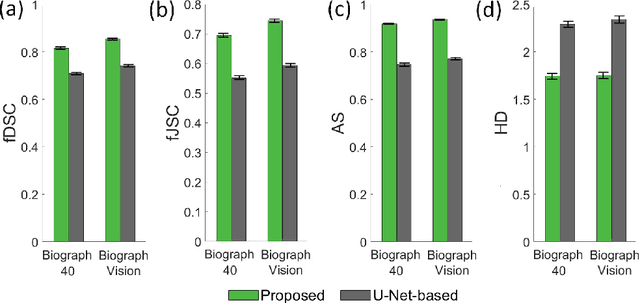


Abstract:Tumor segmentation in oncological PET images is challenging, a major reason being the partial-volume effects that arise from low system resolution and a finite pixel size. The latter results in pixels containing more than one region, also referred to as tissue-fraction effects. Conventional classification-based segmentation approaches are inherently limited in accounting for the tissue-fraction effects. To address this limitation, we pose the segmentation task as an estimation problem. We propose a Bayesian method that estimates the posterior mean of the tumorfraction area within each pixel and uses these estimates to define the segmented tumor boundary. The method was implemented using an autoencoder. Quantitative evaluation of the method was performed using realistic simulation studies conducted in the context of segmenting the primary tumor in PET images of patients with lung cancer. For these studies, a framework was developed to generate clinically realistic simulated PET images. Realism of these images was quantitatively confirmed using a two-alternative-forced-choice study by six trained readers with expertise in reading PET scans. The evaluation studies demonstrated that the proposed segmentation method was accurate, significantly outperformed widely used conventional methods on the tasks of tumor segmentation and estimation of tumor-fraction areas, was relatively insensitive to partial-volume effects, and reliably estimated the ground-truth tumor boundaries. Further, these results were obtained across different clinical-scanner configurations. This proof-of-concept study demonstrates the efficacy of an estimation-based approach to PET segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge