Hae Sol Moon
Fully automated 3D segmentation of dopamine transporter SPECT images using an estimation-based approach
Jan 17, 2021



Abstract:Quantitative measures of uptake in caudate, putamen, and globus pallidus in dopamine transporter (DaT) brain SPECT have potential as biomarkers for the severity of Parkinson disease. Reliable quantification of uptake requires accurate segmentation of these regions. However, segmentation is challenging in DaT SPECT due to partial-volume effects, system noise, physiological variability, and the small size of these regions. To address these challenges, we propose an estimation-based approach to segmentation. This approach estimates the posterior mean of the fractional volume occupied by caudate, putamen, and globus pallidus within each voxel of a 3D SPECT image. The estimate is obtained by minimizing a cost function based on the binary cross-entropy loss between the true and estimated fractional volumes over a population of SPECT images, where the distribution of the true fractional volumes is obtained from magnetic resonance images from clinical populations. The proposed method accounts for both the sources of partial-volume effects in SPECT, namely the limited system resolution and tissue-fraction effects. The method was implemented using an encoder-decoder network and evaluated using realistic clinically guided SPECT simulation studies, where the ground-truth fractional volumes were known. The method significantly outperformed all other considered segmentation methods and yielded accurate segmentation with dice similarity coefficients of ~ 0.80 for all regions. The method was relatively insensitive to changes in voxel size. Further, the method was relatively robust up to +/- 10 degrees of patient head tilt along transaxial, sagittal, and coronal planes. Overall, the results demonstrate the efficacy of the proposed method to yield accurate fully automated segmentation of caudate, putamen, and globus pallidus in 3D DaT-SPECT images.
An estimation-based method to segment PET images
Feb 29, 2020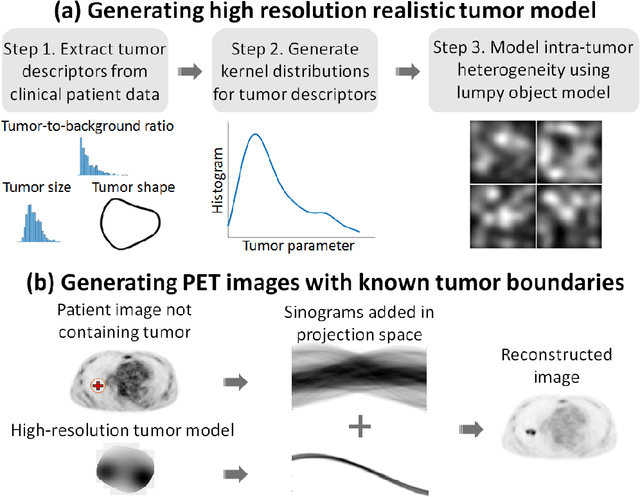
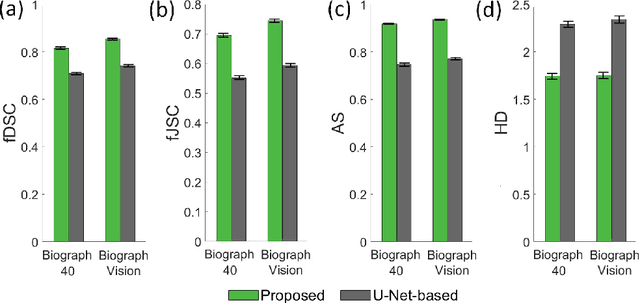


Abstract:Tumor segmentation in oncological PET images is challenging, a major reason being the partial-volume effects that arise from low system resolution and a finite pixel size. The latter results in pixels containing more than one region, also referred to as tissue-fraction effects. Conventional classification-based segmentation approaches are inherently limited in accounting for the tissue-fraction effects. To address this limitation, we pose the segmentation task as an estimation problem. We propose a Bayesian method that estimates the posterior mean of the tumorfraction area within each pixel and uses these estimates to define the segmented tumor boundary. The method was implemented using an autoencoder. Quantitative evaluation of the method was performed using realistic simulation studies conducted in the context of segmenting the primary tumor in PET images of patients with lung cancer. For these studies, a framework was developed to generate clinically realistic simulated PET images. Realism of these images was quantitatively confirmed using a two-alternative-forced-choice study by six trained readers with expertise in reading PET scans. The evaluation studies demonstrated that the proposed segmentation method was accurate, significantly outperformed widely used conventional methods on the tasks of tumor segmentation and estimation of tumor-fraction areas, was relatively insensitive to partial-volume effects, and reliably estimated the ground-truth tumor boundaries. Further, these results were obtained across different clinical-scanner configurations. This proof-of-concept study demonstrates the efficacy of an estimation-based approach to PET segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge