Barry A. Siegel
SPASHT: An image-enhancement method for sparse-view MPI SPECT
Nov 09, 2025Abstract:Single-photon emission computed tomography for myocardial perfusion imaging (MPI SPECT) is a widely used diagnostic tool for coronary artery disease. However, the procedure requires considerable scanning time, leading to patient discomfort and the potential for motion-induced artifacts. Reducing the number of projection views while keeping the time per view unchanged provides a mechanism to shorten the scanning time. However, this approach leads to increased sampling artifacts, higher noise, and hence limited image quality. To address these issues, we propose sparseview SPECT image enhancement (SPASHT), inherently training the algorithm to improve performance on defect-detection tasks. We objectively evaluated SPASHT on the clinical task of detecting perfusion defects in a retrospective clinical study using data from patients who underwent MPI SPECT, where the defects were clinically realistic and synthetically inserted. The study was conducted for different numbers of fewer projection views, including 1/6, 1/3, and 1/2 of the typical projection views for MPI SPECT. Performance on the detection task was quantified using area under the receiver operating characteristic curve (AUC). Images obtained with SPASHT yielded significantly improved AUC compared to those obtained with the sparse-view protocol for all the considered numbers of fewer projection views. To further assess performance, a human observer study on the task of detecting perfusion defects was conducted. Results from the human observer study showed improved detection performance with images reconstructed using SPASHT compared to those from the sparse-view protocol. The results provide evidence of the efficacy of SPASHT in improving the quality of sparse-view MPI SPECT images and motivate further clinical validation.
DEMIST: A deep-learning-based task-specific denoising approach for myocardial perfusion SPECT
Jun 14, 2023Abstract:There is an important need for methods to process myocardial perfusion imaging (MPI) SPECT images acquired at lower radiation dose and/or acquisition time such that the processed images improve observer performance on the clinical task of detecting perfusion defects. To address this need, we build upon concepts from model-observer theory and our understanding of the human visual system to propose a Detection task-specific deep-learning-based approach for denoising MPI SPECT images (DEMIST). The approach, while performing denoising, is designed to preserve features that influence observer performance on detection tasks. We objectively evaluated DEMIST on the task of detecting perfusion defects using a retrospective study with anonymized clinical data in patients who underwent MPI studies across two scanners (N = 338). The evaluation was performed at low-dose levels of 6.25%, 12.5% and 25% and using an anthropomorphic channelized Hotelling observer. Performance was quantified using area under the receiver operating characteristics curve (AUC). Images denoised with DEMIST yielded significantly higher AUC compared to corresponding low-dose images and images denoised with a commonly used task-agnostic DL-based denoising method. Similar results were observed with stratified analysis based on patient sex and defect type. Additionally, DEMIST improved visual fidelity of the low-dose images as quantified using root mean squared error and structural similarity index metric. A mathematical analysis revealed that DEMIST preserved features that assist in detection tasks while improving the noise properties, resulting in improved observer performance. The results provide strong evidence for further clinical evaluation of DEMIST to denoise low-count images in MPI SPECT.
Development and task-based evaluation of a scatter-window projection and deep learning-based transmission-less attenuation compensation method for myocardial perfusion SPECT
Mar 19, 2023Abstract:Attenuation compensation (AC) is beneficial for visual interpretation tasks in single-photon emission computed tomography (SPECT) myocardial perfusion imaging (MPI). However, traditional AC methods require the availability of a transmission scan, most often a CT scan. This approach has the disadvantages of increased radiation dose, increased scanner cost, and the possibility of inaccurate diagnosis in cases of misregistration between the SPECT and CT images. Further, many SPECT systems do not include a CT component. To address these issues, we developed a Scatter-window projection and deep Learning-based AC (SLAC) method to perform AC without a separate transmission scan. To investigate the clinical efficacy of this method, we then objectively evaluated the performance of this method on the clinical task of detecting perfusion defects on MPI in a retrospective study with anonymized clinical SPECT/CT stress MPI images. The proposed method was compared with CT-based AC (CTAC) and no-AC (NAC) methods. Our results showed that the SLAC method yielded an almost overlapping receiver operating characteristic (ROC) plot and a similar area under the ROC (AUC) to the CTAC method on this task. These results demonstrate the capability of the SLAC method for transmission-less AC in SPECT and motivate further clinical evaluation.
Need for Objective Task-based Evaluation of Deep Learning-Based Denoising Methods: A Study in the Context of Myocardial Perfusion SPECT
Mar 16, 2023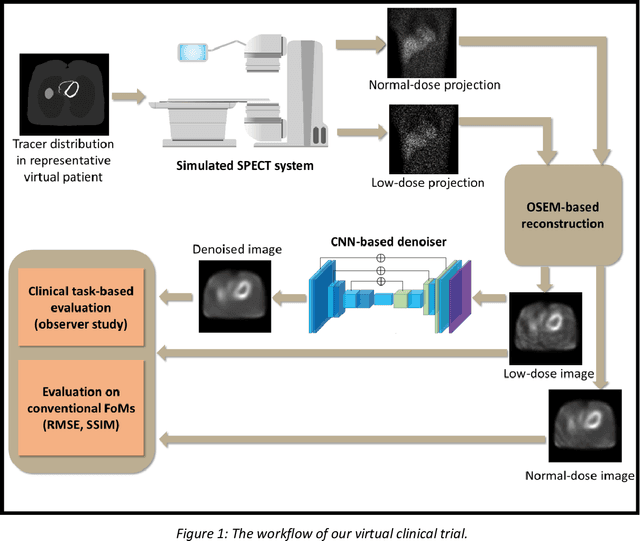
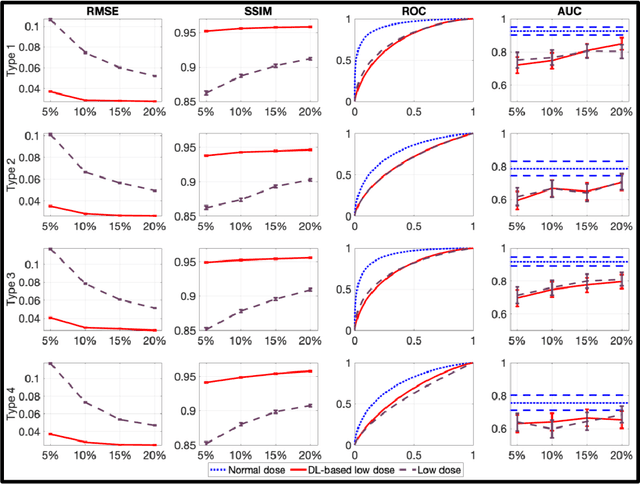
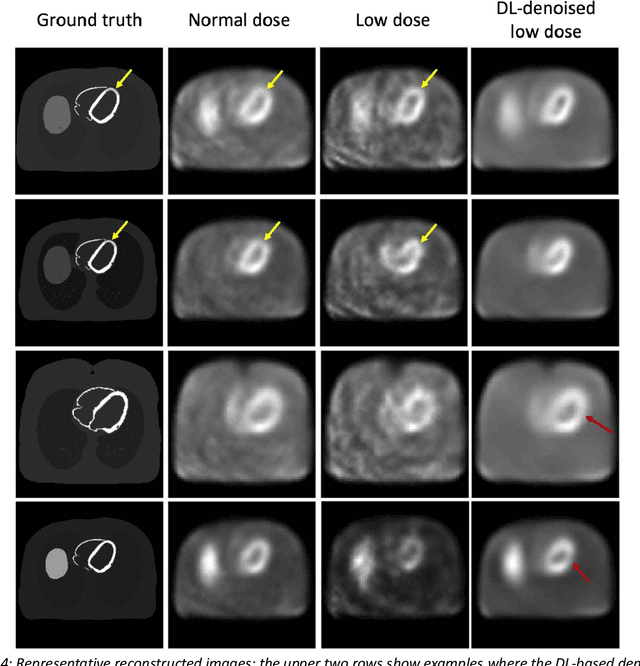
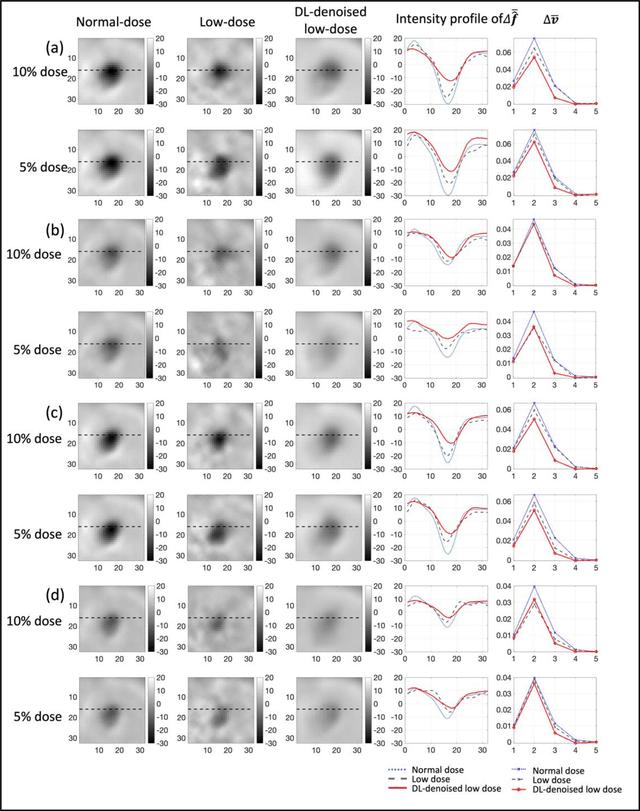
Abstract:Artificial intelligence-based methods have generated substantial interest in nuclear medicine. An area of significant interest has been using deep-learning (DL)-based approaches for denoising images acquired with lower doses, shorter acquisition times, or both. Objective evaluation of these approaches is essential for clinical application. DL-based approaches for denoising nuclear-medicine images have typically been evaluated using fidelity-based figures of merit (FoMs) such as RMSE and SSIM. However, these images are acquired for clinical tasks and thus should be evaluated based on their performance in these tasks. Our objectives were to (1) investigate whether evaluation with these FoMs is consistent with objective clinical-task-based evaluation; (2) provide a theoretical analysis for determining the impact of denoising on signal-detection tasks; (3) demonstrate the utility of virtual clinical trials (VCTs) to evaluate DL-based methods. A VCT to evaluate a DL-based method for denoising myocardial perfusion SPECT (MPS) images was conducted. The impact of DL-based denoising was evaluated using fidelity-based FoMs and AUC, which quantified performance on detecting perfusion defects in MPS images as obtained using a model observer with anthropomorphic channels. Based on fidelity-based FoMs, denoising using the considered DL-based method led to significantly superior performance. However, based on ROC analysis, denoising did not improve, and in fact, often degraded detection-task performance. The results motivate the need for objective task-based evaluation of DL-based denoising approaches. Further, this study shows how VCTs provide a mechanism to conduct such evaluations using VCTs. Finally, our theoretical treatment reveals insights into the reasons for the limited performance of the denoising approach.
A task-specific deep-learning-based denoising approach for myocardial perfusion SPECT
Mar 01, 2023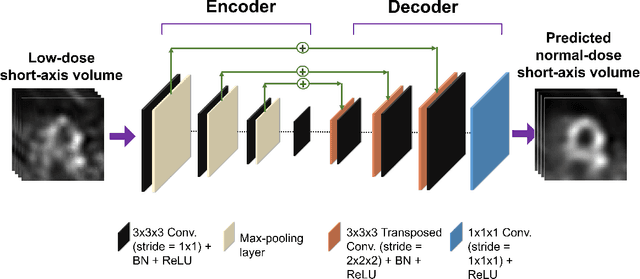
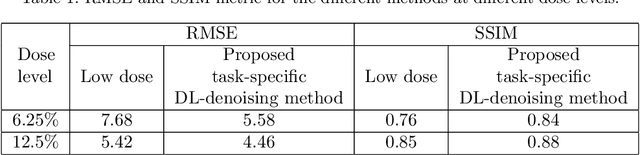
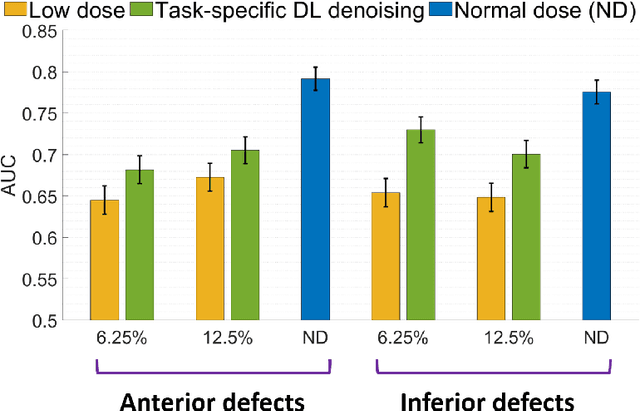
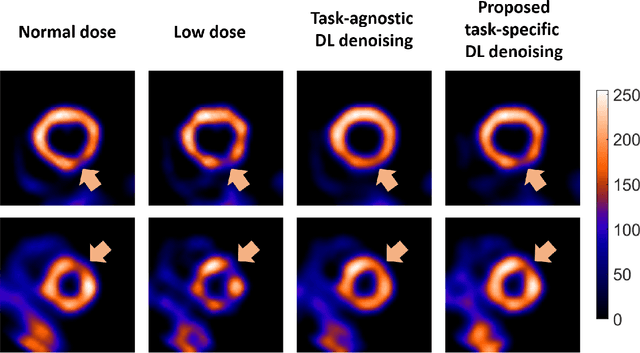
Abstract:Deep-learning (DL)-based methods have shown significant promise in denoising myocardial perfusion SPECT images acquired at low dose. For clinical application of these methods, evaluation on clinical tasks is crucial. Typically, these methods are designed to minimize some fidelity-based criterion between the predicted denoised image and some reference normal-dose image. However, while promising, studies have shown that these methods may have limited impact on the performance of clinical tasks in SPECT. To address this issue, we use concepts from the literature on model observers and our understanding of the human visual system to propose a DL-based denoising approach designed to preserve observer-related information for detection tasks. The proposed method was objectively evaluated on the task of detecting perfusion defect in myocardial perfusion SPECT images using a retrospective study with anonymized clinical data. Our results demonstrate that the proposed method yields improved performance on this detection task compared to using low-dose images. The results show that by preserving task-specific information, DL may provide a mechanism to improve observer performance in low-dose myocardial perfusion SPECT.
No-gold-standard evaluation of quantitative imaging methods in the presence of correlated noise
Mar 03, 2022

Abstract:Objective evaluation of quantitative imaging (QI) methods with patient data is highly desirable, but is hindered by the lack or unreliability of an available gold standard. To address this issue, techniques that can evaluate QI methods without access to a gold standard are being actively developed. These techniques assume that the true and measured values are linearly related by a slope, bias, and Gaussian-distributed noise term, where the noise between measurements made by different methods is independent of each other. However, this noise arises in the process of measuring the same quantitative value, and thus can be correlated. To address this limitation, we propose a no-gold-standard evaluation (NGSE) technique that models this correlated noise by a multi-variate Gaussian distribution parameterized by a covariance matrix. We derive a maximum-likelihood-based approach to estimate the parameters that describe the relationship between the true and measured values, without any knowledge of the true values. We then use the estimated slopes and diagonal elements of the covariance matrix to compute the noise-to-slope ratio (NSR) to rank the QI methods on the basis of precision. The proposed NGSE technique was evaluated with multiple numerical experiments. Our results showed that the technique reliably estimated the NSR values and yielded accurate rankings of the considered methods for ~ 83% of 160 trials. In particular, the technique correctly identified the most precise method for ~ 97% of the trials. Overall, this study demonstrates the efficacy of the NGSE technique to accurately rank different QI methods when the correlated noise is present, and without access to any knowledge of the ground truth. The results motivate further validation of this technique with realistic simulation studies and patient data.
Objective task-based evaluation of artificial intelligence-based medical imaging methods: Framework, strategies and role of the physician
Jul 20, 2021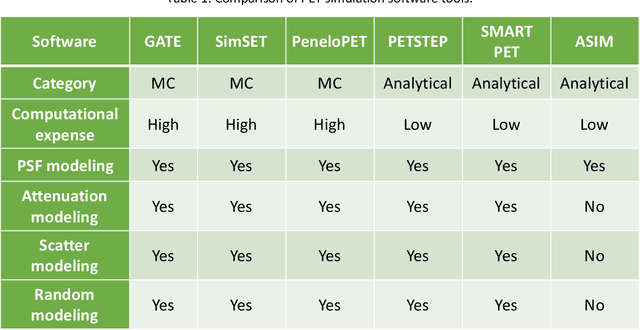
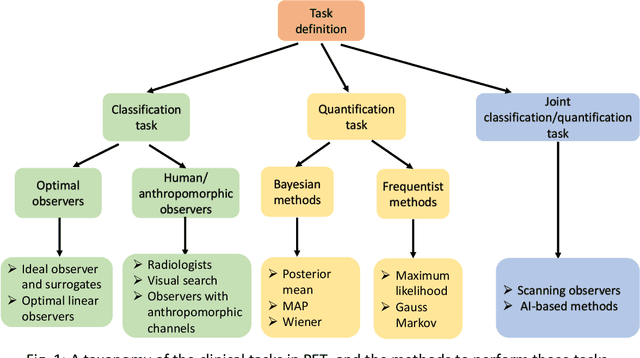
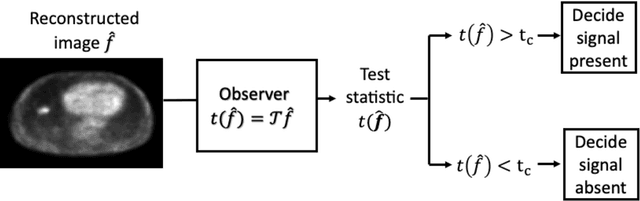
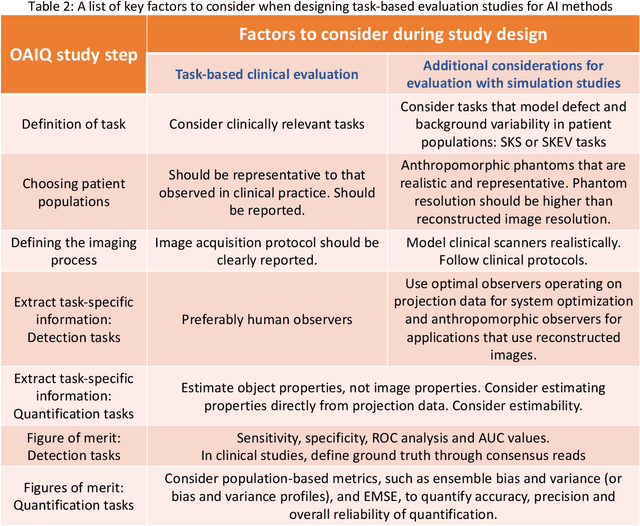
Abstract:Artificial intelligence (AI)-based methods are showing promise in multiple medical-imaging applications. Thus, there is substantial interest in clinical translation of these methods, requiring in turn, that they be evaluated rigorously. In this paper, our goal is to lay out a framework for objective task-based evaluation of AI methods. We will also provide a list of tools available in the literature to conduct this evaluation. Further, we outline the important role of physicians in conducting these evaluation studies. The examples in this paper will be proposed in the context of PET with a focus on neural-network-based methods. However, the framework is also applicable to evaluate other medical-imaging modalities and other types of AI methods.
Observer study-based evaluation of a stochastic and physics-based method to generate oncological PET images
Feb 11, 2021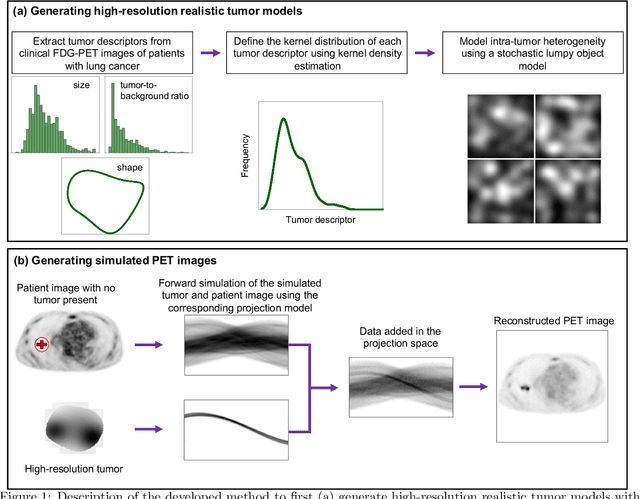


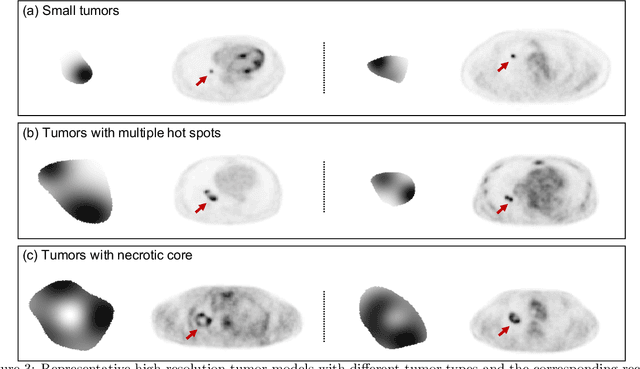
Abstract:Objective evaluation of new and improved methods for PET imaging requires access to images with ground truth, as can be obtained through simulation studies. However, for these studies to be clinically relevant, it is important that the simulated images are clinically realistic. In this study, we develop a stochastic and physics-based method to generate realistic oncological two-dimensional (2-D) PET images, where the ground-truth tumor properties are known. The developed method extends upon a previously proposed approach. The approach captures the observed variabilities in tumor properties from actual patient population. Further, we extend that approach to model intra-tumor heterogeneity using a lumpy object model. To quantitatively evaluate the clinical realism of the simulated images, we conducted a human-observer study. This was a two-alternative forced-choice (2AFC) study with trained readers (five PET physicians and one PET physicist). Our results showed that the readers had an average of ~ 50% accuracy in the 2AFC study. Further, the developed simulation method was able to generate wide varieties of clinically observed tumor types. These results provide evidence for the application of this method to 2-D PET imaging applications, and motivate development of this method to generate 3-D PET images.
An estimation-based method to segment PET images
Feb 29, 2020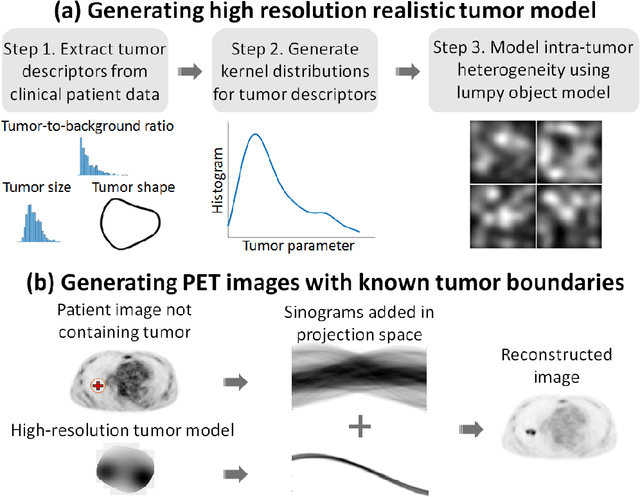
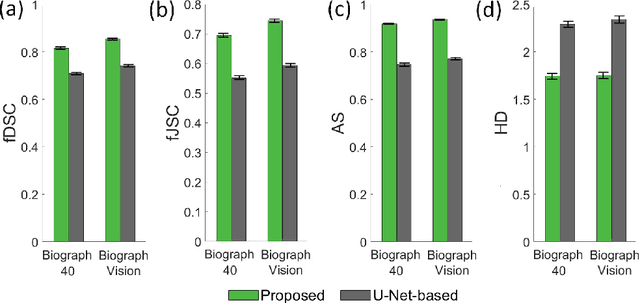


Abstract:Tumor segmentation in oncological PET images is challenging, a major reason being the partial-volume effects that arise from low system resolution and a finite pixel size. The latter results in pixels containing more than one region, also referred to as tissue-fraction effects. Conventional classification-based segmentation approaches are inherently limited in accounting for the tissue-fraction effects. To address this limitation, we pose the segmentation task as an estimation problem. We propose a Bayesian method that estimates the posterior mean of the tumorfraction area within each pixel and uses these estimates to define the segmented tumor boundary. The method was implemented using an autoencoder. Quantitative evaluation of the method was performed using realistic simulation studies conducted in the context of segmenting the primary tumor in PET images of patients with lung cancer. For these studies, a framework was developed to generate clinically realistic simulated PET images. Realism of these images was quantitatively confirmed using a two-alternative-forced-choice study by six trained readers with expertise in reading PET scans. The evaluation studies demonstrated that the proposed segmentation method was accurate, significantly outperformed widely used conventional methods on the tasks of tumor segmentation and estimation of tumor-fraction areas, was relatively insensitive to partial-volume effects, and reliably estimated the ground-truth tumor boundaries. Further, these results were obtained across different clinical-scanner configurations. This proof-of-concept study demonstrates the efficacy of an estimation-based approach to PET segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge