Alexey Chernyavskiy
Self-supervised Physics-based Denoising for Computed Tomography
Nov 01, 2022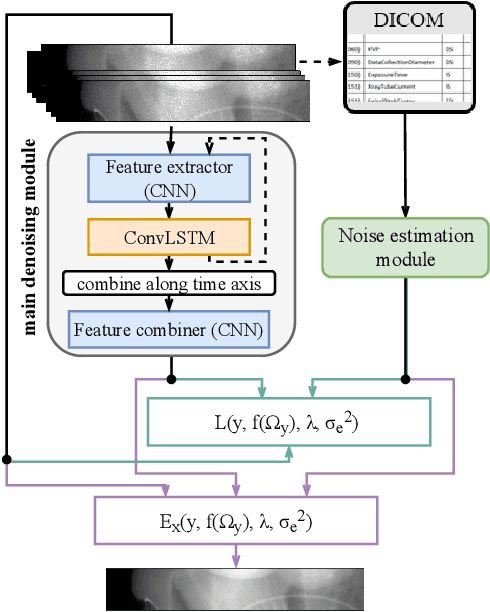


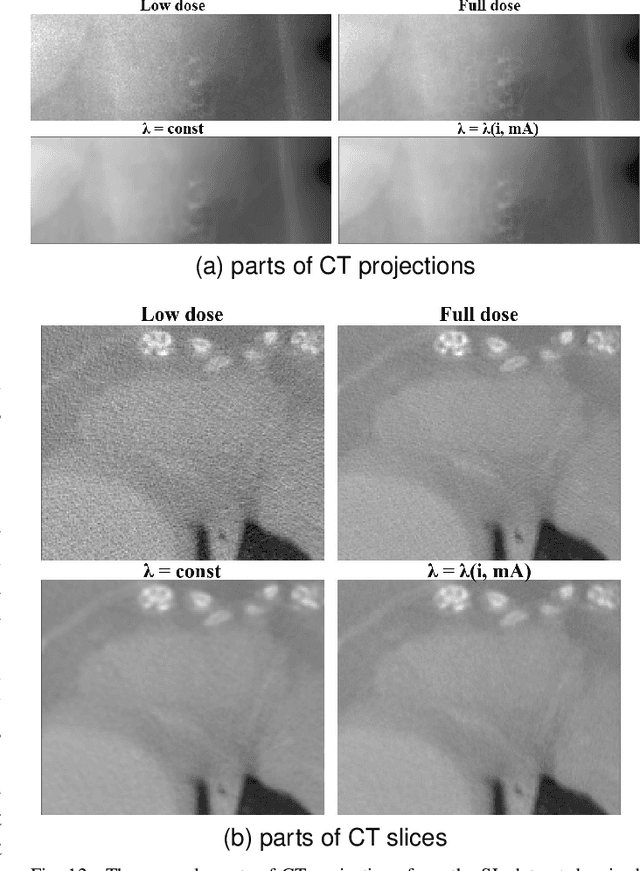
Abstract:Computed Tomography (CT) imposes risk on the patients due to its inherent X-ray radiation, stimulating the development of low-dose CT (LDCT) imaging methods. Lowering the radiation dose reduces the health risks but leads to noisier measurements, which decreases the tissue contrast and causes artifacts in CT images. Ultimately, these issues could affect the perception of medical personnel and could cause misdiagnosis. Modern deep learning noise suppression methods alleviate the challenge but require low-noise-high-noise CT image pairs for training, rarely collected in regular clinical workflows. In this work, we introduce a new self-supervised approach for CT denoising Noise2NoiseTD-ANM that can be trained without the high-dose CT projection ground truth images. Unlike previously proposed self-supervised techniques, the introduced method exploits the connections between the adjacent projections and the actual model of CT noise distribution. Such a combination allows for interpretable no-reference denoising using nothing but the original noisy LDCT projections. Our experiments with LDCT data demonstrate that the proposed method reaches the level of the fully supervised models, sometimes superseding them, easily generalizes to various noise levels, and outperforms state-of-the-art self-supervised denoising algorithms.
Medical image segmentation with imperfect 3D bounding boxes
Aug 06, 2021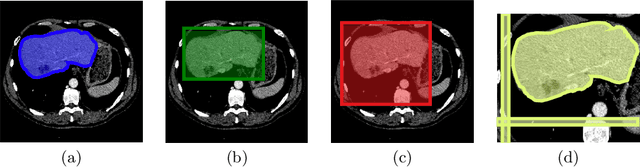



Abstract:The development of high quality medical image segmentation algorithms depends on the availability of large datasets with pixel-level labels. The challenges of collecting such datasets, especially in case of 3D volumes, motivate to develop approaches that can learn from other types of labels that are cheap to obtain, e.g. bounding boxes. We focus on 3D medical images with their corresponding 3D bounding boxes which are considered as series of per-slice non-tight 2D bounding boxes. While current weakly-supervised approaches that use 2D bounding boxes as weak labels can be applied to medical image segmentation, we show that their success is limited in cases when the assumption about the tightness of the bounding boxes breaks. We propose a new bounding box correction framework which is trained on a small set of pixel-level annotations to improve the tightness of a larger set of non-tight bounding box annotations. The effectiveness of our solution is demonstrated by evaluating a known weakly-supervised segmentation approach with and without the proposed bounding box correction algorithm. When the tightness is improved by our solution, the results of the weakly-supervised segmentation become much closer to those of the fully-supervised one.
Anatomy of Domain Shift Impact on U-Net Layers in MRI Segmentation
Jul 10, 2021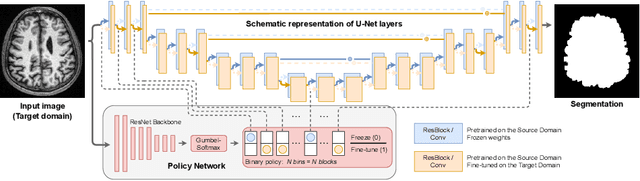
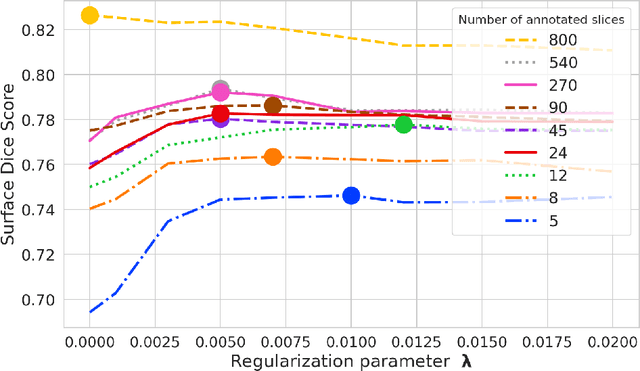
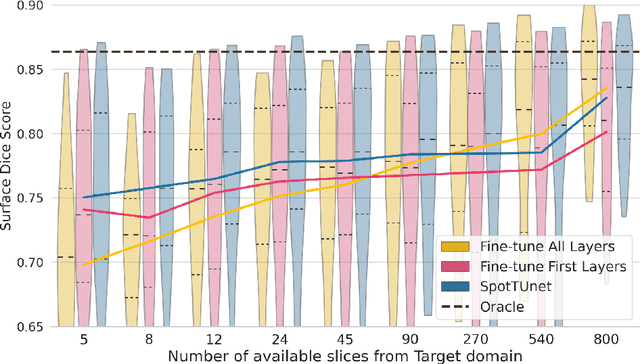
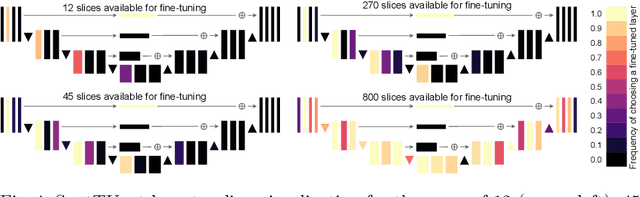
Abstract:Domain Adaptation (DA) methods are widely used in medical image segmentation tasks to tackle the problem of differently distributed train (source) and test (target) data. We consider the supervised DA task with a limited number of annotated samples from the target domain. It corresponds to one of the most relevant clinical setups: building a sufficiently accurate model on the minimum possible amount of annotated data. Existing methods mostly fine-tune specific layers of the pretrained Convolutional Neural Network (CNN). However, there is no consensus on which layers are better to fine-tune, e.g. the first layers for images with low-level domain shift or the deeper layers for images with high-level domain shift. To this end, we propose SpotTUnet - a CNN architecture that automatically chooses the layers which should be optimally fine-tuned. More specifically, on the target domain, our method additionally learns the policy that indicates whether a specific layer should be fine-tuned or reused from the pretrained network. We show that our method performs at the same level as the best of the nonflexible fine-tuning methods even under the extreme scarcity of annotated data. Secondly, we show that SpotTUnet policy provides a layer-wise visualization of the domain shift impact on the network, which could be further used to develop robust domain generalization methods. In order to extensively evaluate SpotTUnet performance, we use a publicly available dataset of brain MR images (CC359), characterized by explicit domain shift. We release a reproducible experimental pipeline.
Uncertainty-based method for improving poorly labeled segmentation datasets
Feb 16, 2021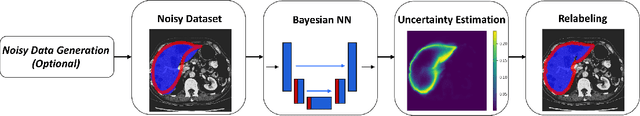
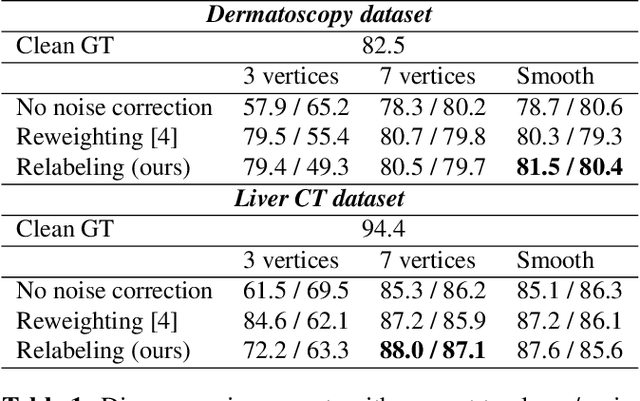
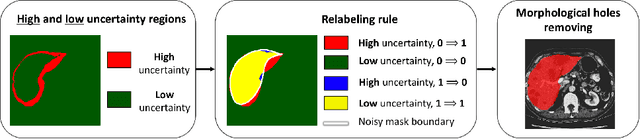
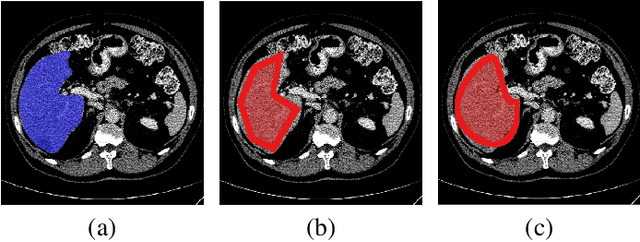
Abstract:The success of modern deep learning algorithms for image segmentation heavily depends on the availability of large datasets with clean pixel-level annotations (masks), where the objects of interest are accurately delineated. Lack of time and expertise during data annotation leads to incorrect boundaries and label noise. It is known that deep convolutional neural networks (DCNNs) can memorize even completely random labels, resulting in poor accuracy. We propose a framework to train binary segmentation DCNNs using sets of unreliable pixel-level annotations. Erroneously labeled pixels are identified based on the estimated aleatoric uncertainty of the segmentation and are relabeled to the true value.
No-reference denoising of low-dose CT projections
Feb 03, 2021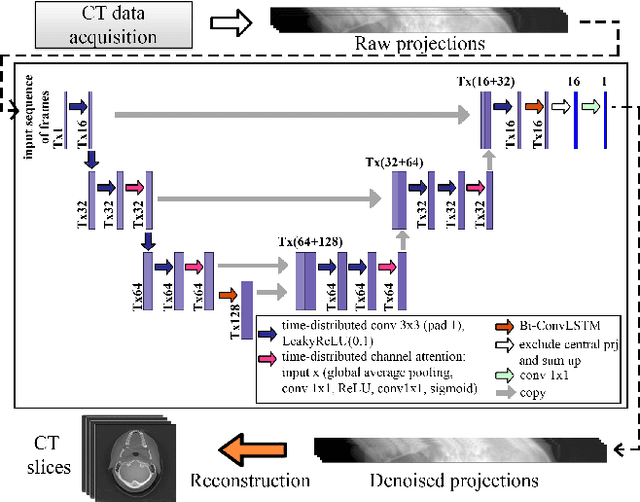
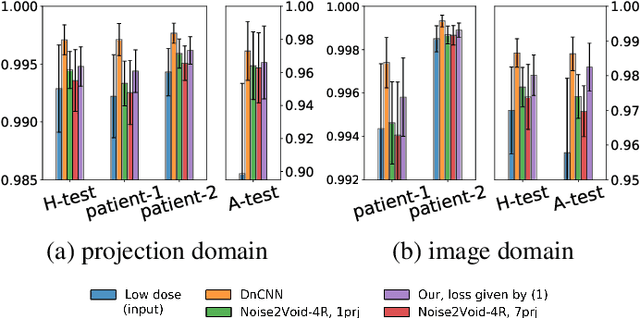
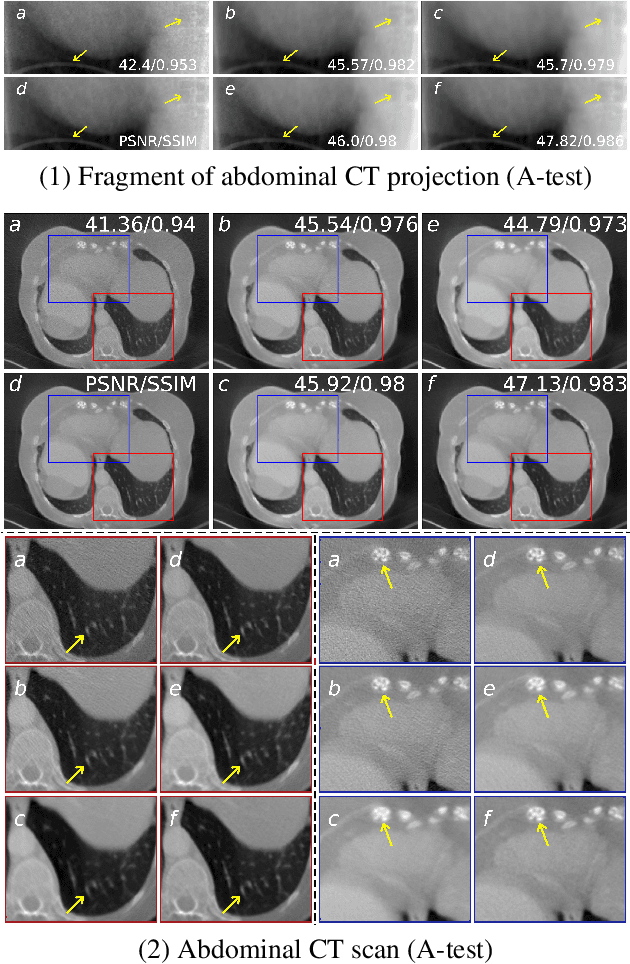
Abstract:Low-dose computed tomography (LDCT) became a clear trend in radiology with an aspiration to refrain from delivering excessive X-ray radiation to the patients. The reduction of the radiation dose decreases the risks to the patients but raises the noise level, affecting the quality of the images and their ultimate diagnostic value. One mitigation option is to consider pairs of low-dose and high-dose CT projections to train a denoising model using deep learning algorithms; however, such pairs are rarely available in practice. In this paper, we present a new self-supervised method for CT denoising. Unlike existing self-supervised approaches, the proposed method requires only noisy CT projections and exploits the connections between adjacent images. The experiments carried out on an LDCT dataset demonstrate that our method is almost as accurate as the supervised approach, while also outperforming the considered self-supervised denoising methods.
First U-Net Layers Contain More Domain Specific Information Than The Last Ones
Aug 17, 2020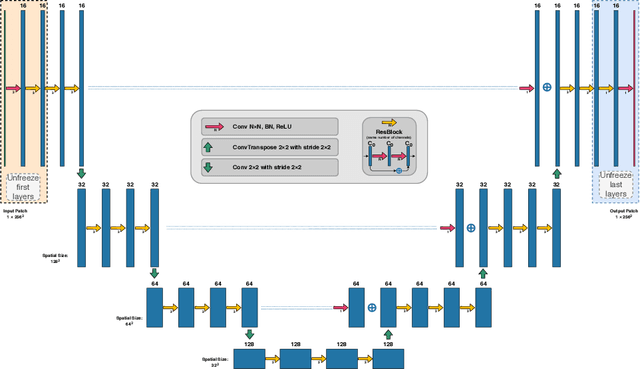
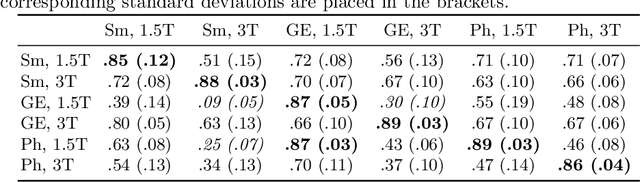
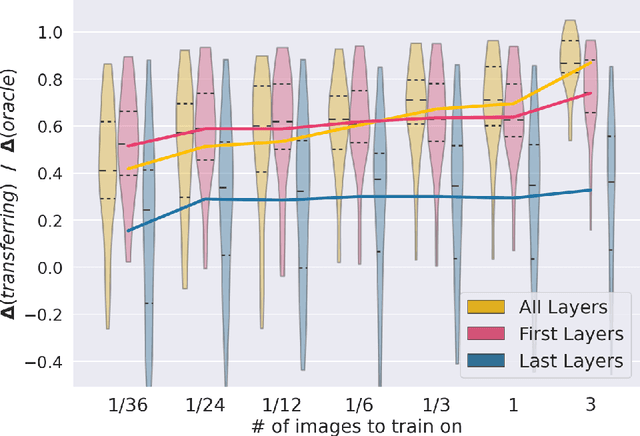
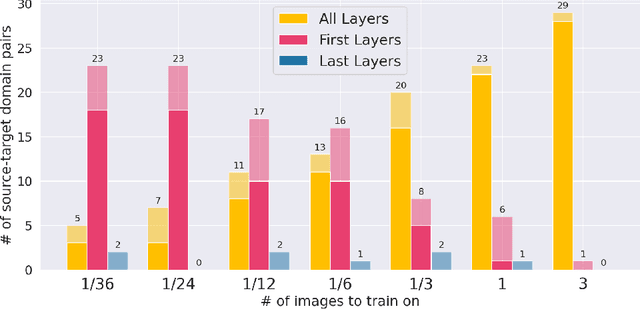
Abstract:MRI scans appearance significantly depends on scanning protocols and, consequently, the data-collection institution. These variations between clinical sites result in dramatic drops of CNN segmentation quality on unseen domains. Many of the recently proposed MRI domain adaptation methods operate with the last CNN layers to suppress domain shift. At the same time, the core manifestation of MRI variability is a considerable diversity of image intensities. We hypothesize that these differences can be eliminated by modifying the first layers rather than the last ones. To validate this simple idea, we conducted a set of experiments with brain MRI scans from six domains. Our results demonstrate that 1) domain-shift may deteriorate the quality even for a simple brain extraction segmentation task (surface Dice Score drops from 0.85-0.89 even to 0.09); 2) fine-tuning of the first layers significantly outperforms fine-tuning of the last layers in almost all supervised domain adaptation setups. Moreover, fine-tuning of the first layers is a better strategy than fine-tuning of the whole network, if the amount of annotated data from the new domain is strictly limited.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge