Dong Nie
Seeing the Forest and the Trees: Query-Aware Tokenizer for Long-Video Multimodal Language Models
Nov 14, 2025Abstract:Despite the recent advances in the video understanding ability of multimodal large language models (MLLMs), long video understanding remains a challenge. One of the main issues is that the number of vision tokens grows linearly with video length, which causes an explosion in attention cost, memory, and latency. To solve this challenge, we present Query-aware Token Selector (\textbf{QTSplus}), a lightweight yet powerful visual token selection module that serves as an information gate between the vision encoder and LLMs. Given a text query and video tokens, QTSplus dynamically selects the most important visual evidence for the input text query by (i) scoring visual tokens via cross-attention, (ii) \emph{predicting} an instance-specific retention budget based on the complexity of the query, and (iii) \emph{selecting} Top-$n$ tokens with a differentiable straight-through estimator during training and a hard gate at inference. Furthermore, a small re-encoder preserves temporal order using absolute time information, enabling second-level localization while maintaining global coverage. Integrated into Qwen2.5-VL, QTSplus compresses the vision stream by up to \textbf{89\%} and reduces end-to-end latency by \textbf{28\%} on long videos. The evaluation on eight long video understanding benchmarks shows near-parity accuracy overall when compared with the original Qwen models and outperforms the original model by \textbf{+20.5} and \textbf{+5.6} points respectively on TempCompass direction and order accuracies. These results show that QTSplus is an effective, general mechanism for scaling MLLMs to real-world long-video scenarios while preserving task-relevant evidence. We will make all code, data, and trained models' weights publicly available.
ViT3D Alignment of LLaMA3: 3D Medical Image Report Generation
Oct 11, 2024



Abstract:Automatic medical report generation (MRG), which aims to produce detailed text reports from medical images, has emerged as a critical task in this domain. MRG systems can enhance radiological workflows by reducing the time and effort required for report writing, thereby improving diagnostic efficiency. In this work, we present a novel approach for automatic MRG utilizing a multimodal large language model. Specifically, we employed the 3D Vision Transformer (ViT3D) image encoder introduced from M3D-CLIP to process 3D scans and use the Asclepius-Llama3-8B as the language model to generate the text reports by auto-regressive decoding. The experiment shows our model achieved an average Green score of 0.3 on the MRG task validation set and an average accuracy of 0.61 on the visual question answering (VQA) task validation set, outperforming the baseline model. Our approach demonstrates the effectiveness of the ViT3D alignment of LLaMA3 for automatic MRG and VQA tasks by tuning the model on a small dataset.
The Devil is in the Upsampling: Architectural Decisions Made Simpler for Denoising with Deep Image Prior
Apr 22, 2023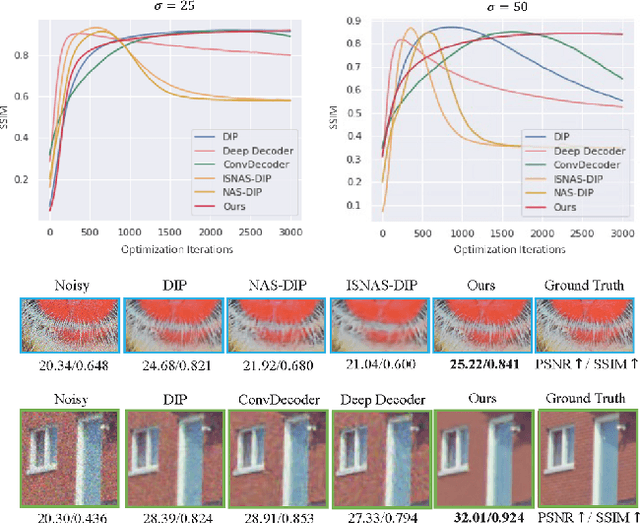
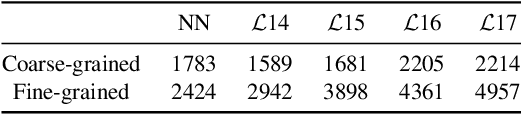
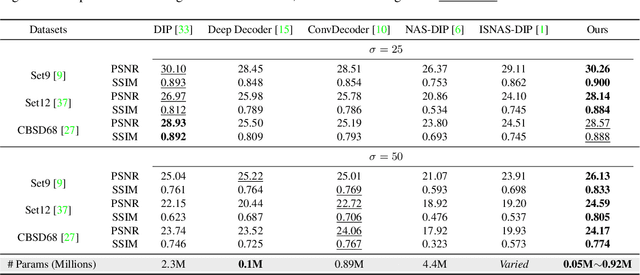
Abstract:Deep Image Prior (DIP) shows that some network architectures naturally bias towards smooth images and resist noises, a phenomenon known as spectral bias. Image denoising is an immediate application of this property. Although DIP has removed the requirement of large training sets, it still presents two practical challenges for denoising: architectural design and noise-fitting, which are often intertwined. Existing methods mostly handcraft or search for the architecture from a large design space, due to the lack of understanding on how the architectural choice corresponds to the image. In this study, we analyze from a frequency perspective to demonstrate that the unlearnt upsampling is the main driving force behind the denoising phenomenon in DIP. This finding then leads to strategies for estimating a suitable architecture for every image without a laborious search. Extensive experiments show that the estimated architectures denoise and preserve the textural details better than current methods with up to 95% fewer parameters. The under-parameterized nature also makes them especially robust to a higher level of noise.
Multi-Label Clinical Time-Series Generation via Conditional GAN
Apr 10, 2022



Abstract:With wide applications of electronic health records (EHR), deep learning methods have been adopted to analyze EHR data on various tasks such as representation learning, clinical event prediction, and phenotyping. However, due to privacy constraints, limited access to EHR becomes a bottleneck for deep learning research. Recently, generative adversarial networks (GANs) have been successful in generating EHR data. However, there are still challenges in high-quality EHR generation, including generating time-series EHR and uncommon diseases given imbalanced datasets. In this work, we propose a Multi-label Time-series GAN (MTGAN) to generate EHR data and simultaneously improve the quality of uncommon disease generation. The generator of MTGAN uses a gated recurrent unit (GRU) with a smooth conditional matrix to generate sequences and uncommon diseases. The critic gives scores using Wasserstein distance to recognize real samples from synthetic samples by considering both data and temporal features. We also propose a training strategy to calculate temporal features for real data and stabilize GAN training. Furthermore, we design multiple statistical metrics and prediction tasks to evaluate the generated data. Experimental results demonstrate the quality of the synthetic data and the effectiveness of MTGAN in generating realistic sequential EHR data, especially for uncommon diseases.
Fast T2w/FLAIR MRI Acquisition by Optimal Sampling of Information Complementary to Pre-acquired T1w MRI
Nov 11, 2021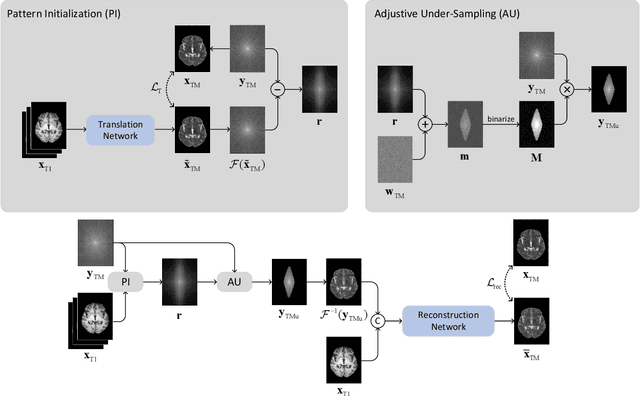
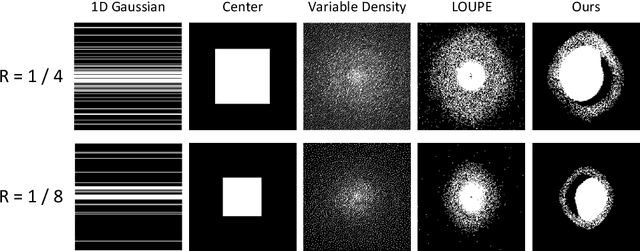

Abstract:Recent studies on T1-assisted MRI reconstruction for under-sampled images of other modalities have demonstrated the potential of further accelerating MRI acquisition of other modalities. Most of the state-of-the-art approaches have achieved improvement through the development of network architectures for fixed under-sampling patterns, without fully exploiting the complementary information between modalities. Although existing under-sampling pattern learning algorithms can be simply modified to allow the fully-sampled T1-weighted MR image to assist the pattern learning, no significant improvement on the reconstruction task can be achieved. To this end, we propose an iterative framework to optimize the under-sampling pattern for MRI acquisition of another modality that can complement the fully-sampled T1-weighted MR image at different under-sampling factors, while jointly optimizing the T1-assisted MRI reconstruction model. Specifically, our proposed method exploits the difference of latent information between the two modalities for determining the sampling patterns that can maximize the assistance power of T1-weighted MR image in improving the MRI reconstruction. We have demonstrated superior performance of our learned under-sampling patterns on a public dataset, compared to commonly used under-sampling patterns and state-of-the-art methods that can jointly optimize both the reconstruction network and the under-sampling pattern, up to 8-fold under-sampling factor.
Automated Generation of Accurate \& Fluent Medical X-ray Reports
Aug 27, 2021



Abstract:Our paper focuses on automating the generation of medical reports from chest X-ray image inputs, a critical yet time-consuming task for radiologists. Unlike existing medical re-port generation efforts that tend to produce human-readable reports, we aim to generate medical reports that are both fluent and clinically accurate. This is achieved by our fully differentiable and end-to-end paradigm containing three complementary modules: taking the chest X-ray images and clinical his-tory document of patients as inputs, our classification module produces an internal check-list of disease-related topics, referred to as enriched disease embedding; the embedding representation is then passed to our transformer-based generator, giving rise to the medical reports; meanwhile, our generator also pro-duces the weighted embedding representation, which is fed to our interpreter to ensure consistency with respect to disease-related topics.Our approach achieved promising results on commonly-used metrics concerning language fluency and clinical accuracy. Moreover, noticeable performance gains are consistently ob-served when additional input information is available, such as the clinical document and extra scans of different views.
HF-UNet: Learning Hierarchically Inter-Task Relevance in Multi-Task U-Net for Accurate Prostate Segmentation
May 23, 2020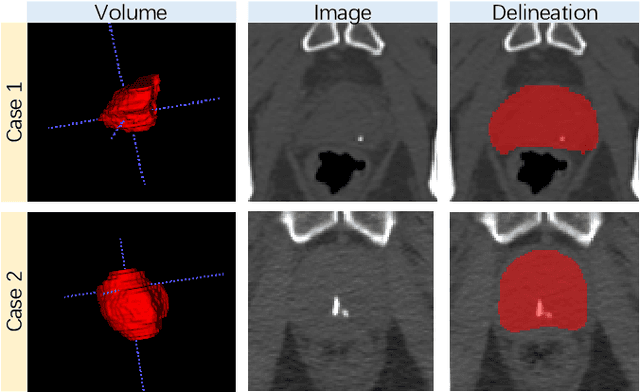
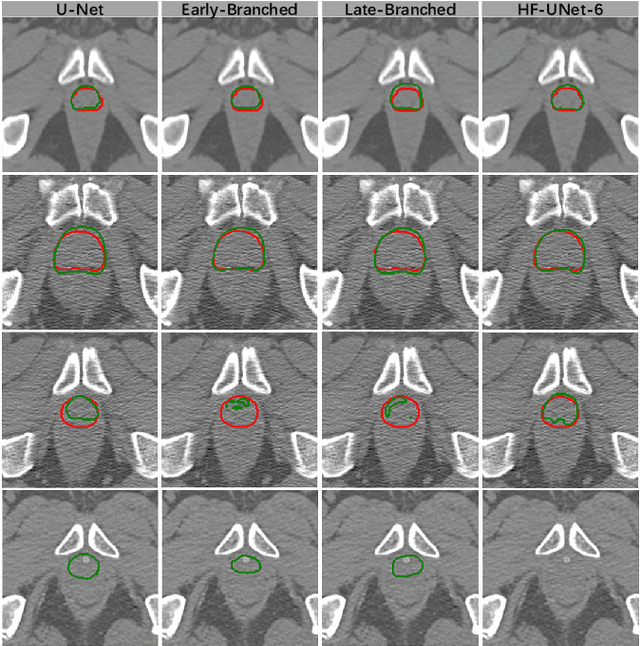
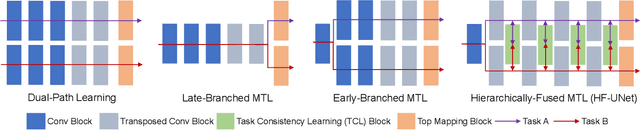
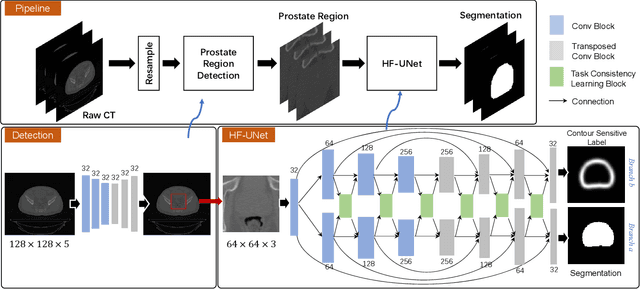
Abstract:Accurate segmentation of the prostate is a key step in external beam radiation therapy treatments. In this paper, we tackle the challenging task of prostate segmentation in CT images by a two-stage network with 1) the first stage to fast localize, and 2) the second stage to accurately segment the prostate. To precisely segment the prostate in the second stage, we formulate prostate segmentation into a multi-task learning framework, which includes a main task to segment the prostate, and an auxiliary task to delineate the prostate boundary. Here, the second task is applied to provide additional guidance of unclear prostate boundary in CT images. Besides, the conventional multi-task deep networks typically share most of the parameters (i.e., feature representations) across all tasks, which may limit their data fitting ability, as the specificities of different tasks are inevitably ignored. By contrast, we solve them by a hierarchically-fused U-Net structure, namely HF-UNet. The HF-UNet has two complementary branches for two tasks, with the novel proposed attention-based task consistency learning block to communicate at each level between the two decoding branches. Therefore, HF-UNet endows the ability to learn hierarchically the shared representations for different tasks, and preserve the specificities of learned representations for different tasks simultaneously. We did extensive evaluations of the proposed method on a large planning CT image dataset, including images acquired from 339 patients. The experimental results show HF-UNet outperforms the conventional multi-task network architectures and the state-of-the-art methods.
Hybrid Graph Neural Networks for Crowd Counting
Jan 31, 2020



Abstract:Crowd counting is an important yet challenging task due to the large scale and density variation. Recent investigations have shown that distilling rich relations among multi-scale features and exploiting useful information from the auxiliary task, i.e., localization, are vital for this task. Nevertheless, how to comprehensively leverage these relations within a unified network architecture is still a challenging problem. In this paper, we present a novel network structure called Hybrid Graph Neural Network (HyGnn) which targets to relieve the problem by interweaving the multi-scale features for crowd density as well as its auxiliary task (localization) together and performing joint reasoning over a graph. Specifically, HyGnn integrates a hybrid graph to jointly represent the task-specific feature maps of different scales as nodes, and two types of relations as edges:(i) multi-scale relations for capturing the feature dependencies across scales and (ii) mutual beneficial relations building bridges for the cooperation between counting and localization. Thus, through message passing, HyGnn can distill rich relations between the nodes to obtain more powerful representations, leading to robust and accurate results. Our HyGnn performs significantly well on four challenging datasets: ShanghaiTech Part A, ShanghaiTech Part B, UCF_CC_50 and UCF_QNRF, outperforming the state-of-the-art approaches by a large margin.
Dual Adversarial Learning with Attention Mechanism for Fine-grained Medical Image Synthesis
Jul 07, 2019
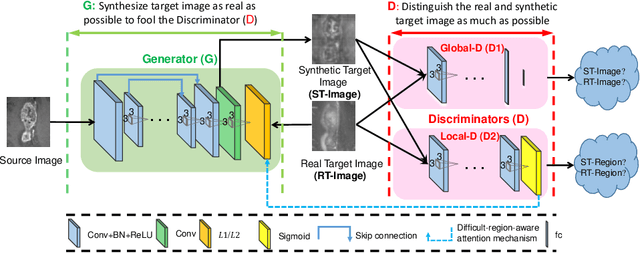

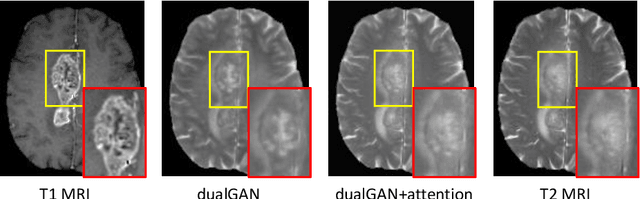
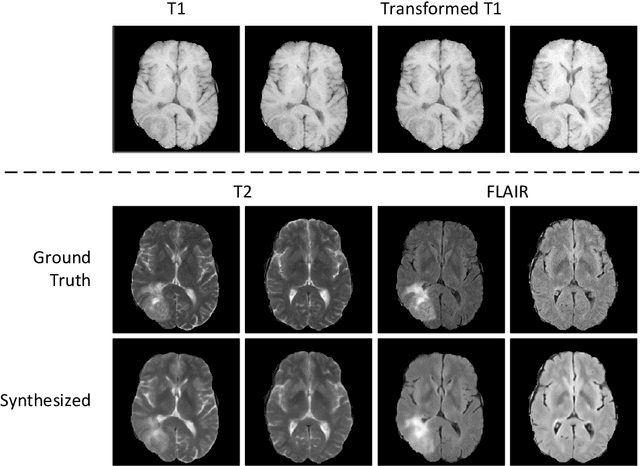
Abstract:Medical imaging plays a critical role in various clinical applications. However, due to multiple considerations such as cost and risk, the acquisition of certain image modalities could be limited. To address this issue, many cross-modality medical image synthesis methods have been proposed. However, the current methods cannot well model the hard-to-synthesis regions (e.g., tumor or lesion regions). To address this issue, we propose a simple but effective strategy, that is, we propose a dual-discriminator (dual-D) adversarial learning system, in which, a global-D is used to make an overall evaluation for the synthetic image, and a local-D is proposed to densely evaluate the local regions of the synthetic image. More importantly, we build an adversarial attention mechanism which targets at better modeling hard-to-synthesize regions (e.g., tumor or lesion regions) based on the local-D. Experimental results show the robustness and accuracy of our method in synthesizing fine-grained target images from the corresponding source images. In particular, we evaluate our method on two datasets, i.e., to address the tasks of generating T2 MRI from T1 MRI for the brain tumor images and generating MRI from CT. Our method outperforms the state-of-the-art methods under comparison in all datasets and tasks. And the proposed difficult-region-aware attention mechanism is also proved to be able to help generate more realistic images, especially for the hard-to-synthesize regions.
Semantic-guided Encoder Feature Learning for Blurry Boundary Delineation
Jun 10, 2019



Abstract:Encoder-decoder architectures are widely adopted for medical image segmentation tasks. With the lateral skip connection, the models can obtain and fuse both semantic and resolution information in deep layers to achieve more accurate segmentation performance. However, in many applications (e.g., blurry boundary images), these models often cannot precisely locate complex boundaries and segment tiny isolated parts. To solve this challenging problem, we firstly analyze why simple skip connections are not enough to help accurately locate indistinct boundaries and argue that it is due to the fuzzy information in the skip connection provided in the encoder layers. Then we propose a semantic-guided encoder feature learning strategy to learn both high resolution and rich semantic encoder features so that we can more accurately locate the blurry boundaries, which can also enhance the network by selectively learning discriminative features. Besides, we further propose a soft contour constraint mechanism to model the blurry boundary detection. Experimental results on real clinical datasets show that our proposed method can achieve state-of-the-art segmentation accuracy, especially for the blurry regions. Further analysis also indicates that our proposed network components indeed contribute to the improvement of performance. Experiments on additional datasets validate the generalization ability of our proposed method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge