Junfeng Zhang
One-step Structure Prediction and Screening for Protein-Ligand Complexes using Multi-Task Geometric Deep Learning
Aug 21, 2024



Abstract:Understanding the structure of the protein-ligand complex is crucial to drug development. Existing virtual structure measurement and screening methods are dominated by docking and its derived methods combined with deep learning. However, the sampling and scoring methodology have largely restricted the accuracy and efficiency. Here, we show that these two fundamental tasks can be accurately tackled with a single model, namely LigPose, based on multi-task geometric deep learning. By representing the ligand and the protein pair as a graph, LigPose directly optimizes the three-dimensional structure of the complex, with the learning of binding strength and atomic interactions as auxiliary tasks, enabling its one-step prediction ability without docking tools. Extensive experiments show LigPose achieved state-of-the-art performance on major tasks in drug research. Its considerable improvements indicate a promising paradigm of AI-based pipeline for drug development.
Transformers in Medical Image Analysis: A Review
Feb 24, 2022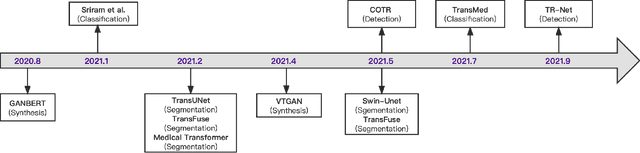
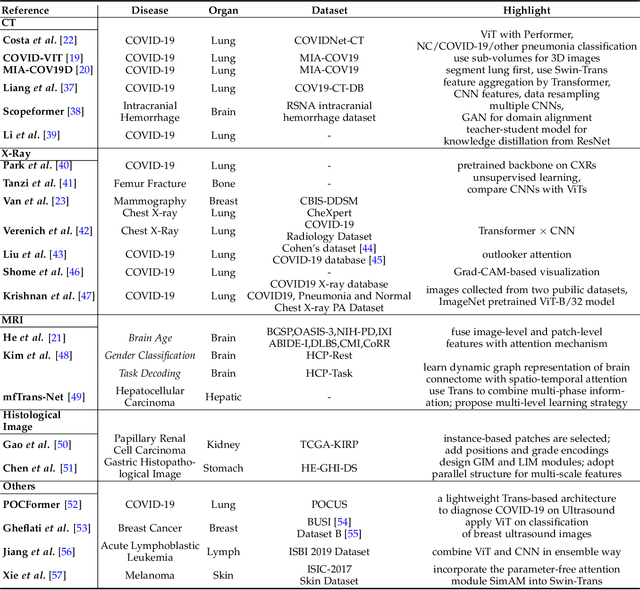
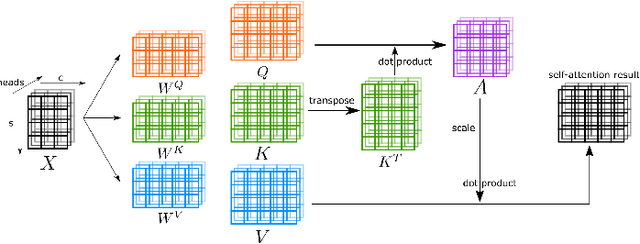
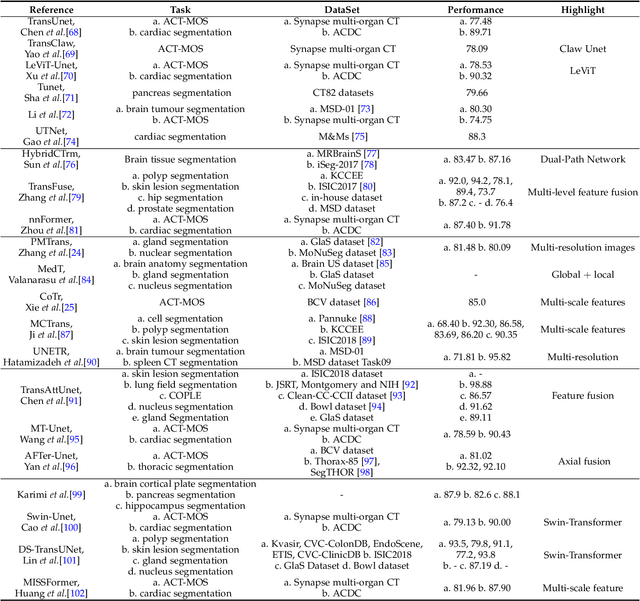
Abstract:Transformers have dominated the field of natural language processing, and recently impacted the computer vision area. In the field of medical image analysis, Transformers have also been successfully applied to full-stack clinical applications, including image synthesis/reconstruction, registration, segmentation, detection, and diagnosis. Our paper presents both a position paper and a primer, promoting awareness and application of Transformers in the field of medical image analysis. Specifically, we first overview the core concepts of the attention mechanism built into Transformers and other basic components. Second, we give a new taxonomy of various Transformer architectures tailored for medical image applications and discuss their limitations. Within this review, we investigate key challenges revolving around the use of Transformers in different learning paradigms, improving the model efficiency, and their coupling with other techniques. We hope this review can give a comprehensive picture of Transformers to the readers in the field of medical image analysis.
Cross-Modality Brain Tumor Segmentation via Bidirectional Global-to-Local Unsupervised Domain Adaptation
May 17, 2021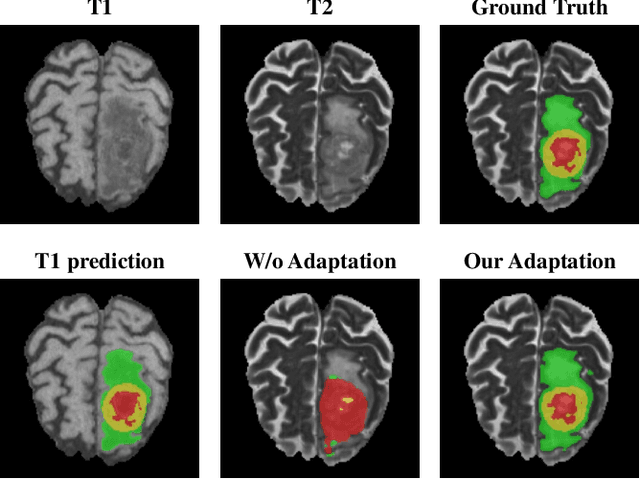
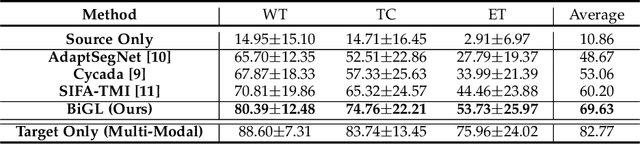
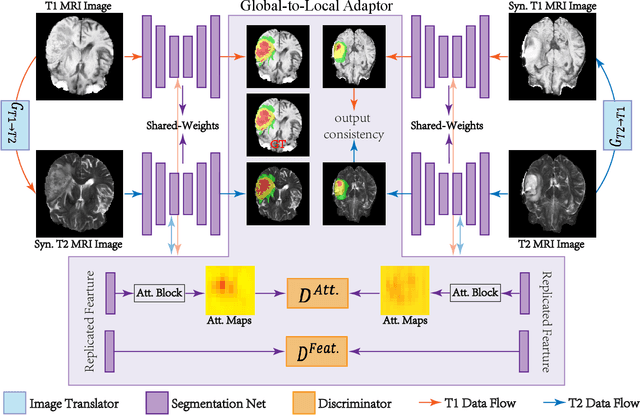
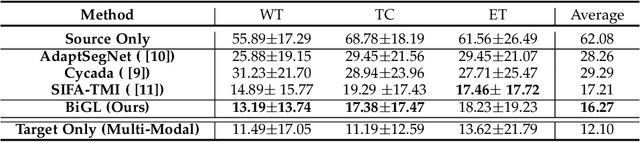
Abstract:Accurate segmentation of brain tumors from multi-modal Magnetic Resonance (MR) images is essential in brain tumor diagnosis and treatment. However, due to the existence of domain shifts among different modalities, the performance of networks decreases dramatically when training on one modality and performing on another, e.g., train on T1 image while performing on T2 image, which is often required in clinical applications. This also prohibits a network from being trained on labeled data and then transferred to unlabeled data from a different domain. To overcome this, unsupervised domain adaptation (UDA) methods provide effective solutions to alleviate the domain shift between labeled source data and unlabeled target data. In this paper, we propose a novel Bidirectional Global-to-Local (BiGL) adaptation framework under a UDA scheme. Specifically, a bidirectional image synthesis and segmentation module is proposed to segment the brain tumor using the intermediate data distributions generated for the two domains, which includes an image-to-image translator and a shared-weighted segmentation network. Further, a global-to-local consistency learning module is proposed to build robust representation alignments in an integrated way. Extensive experiments on a multi-modal brain MR benchmark dataset demonstrate that the proposed method outperforms several state-of-the-art unsupervised domain adaptation methods by a large margin, while a comprehensive ablation study validates the effectiveness of each key component. The implementation code of our method will be released at \url{https://github.com/KeleiHe/BiGL}.
Synergistic Learning of Lung Lobe Segmentation and Hierarchical Multi-Instance Classification for Automated Severity Assessment of COVID-19 in CT Images
May 24, 2020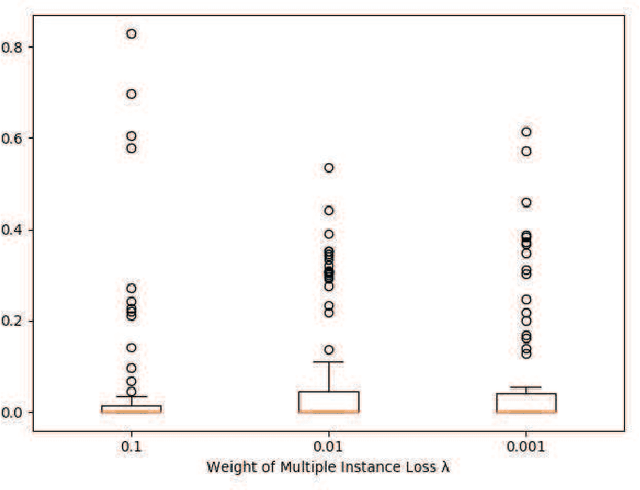
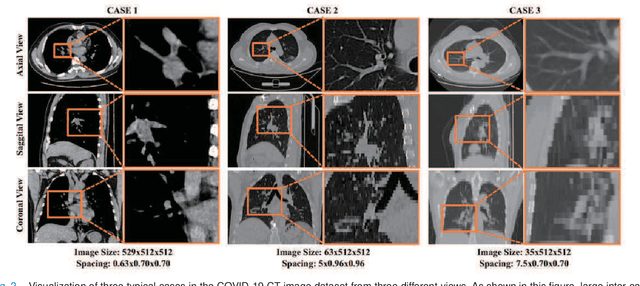
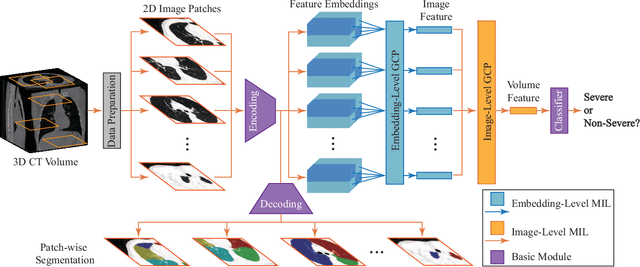
Abstract:Understanding chest CT imaging of the coronavirus disease 2019 (COVID-19) will help detect infections early and assess the disease progression. Especially, automated severity assessment of COVID-19 in CT images plays an essential role in identifying cases that are in great need of intensive clinical care. However, it is often challenging to accurately assess the severity of this disease in CT images, due to variable infection regions in the lungs, similar imaging biomarkers, and large inter-case variations. To this end, we propose a synergistic learning framework for automated severity assessment of COVID-19 in 3D CT images, by jointly performing lung lobe segmentation and multi-instance classification. Considering that only a few infection regions in a CT image are related to the severity assessment, we first represent each input image by a bag that contains a set of 2D image patches (with each cropped from a specific slice). A multi-task multi-instance deep network (called M$^2$UNet) is then developed to assess the severity of COVID-19 patients and also segment the lung lobe simultaneously. Our M$^2$UNet consists of a patch-level encoder, a segmentation sub-network for lung lobe segmentation, and a classification sub-network for severity assessment (with a unique hierarchical multi-instance learning strategy). Here, the context information provided by segmentation can be implicitly employed to improve the performance of severity assessment. Extensive experiments were performed on a real COVID-19 CT image dataset consisting of 666 chest CT images, with results suggesting the effectiveness of our proposed method compared to several state-of-the-art methods.
HF-UNet: Learning Hierarchically Inter-Task Relevance in Multi-Task U-Net for Accurate Prostate Segmentation
May 23, 2020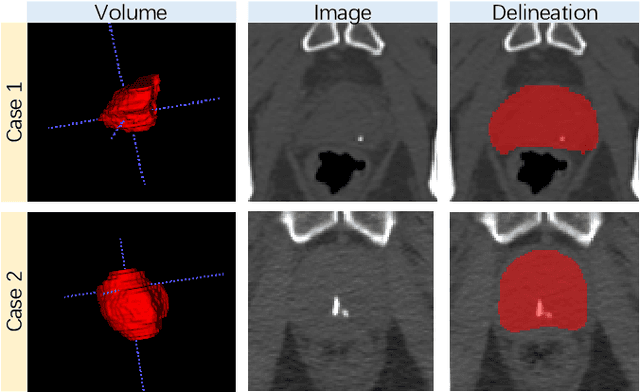
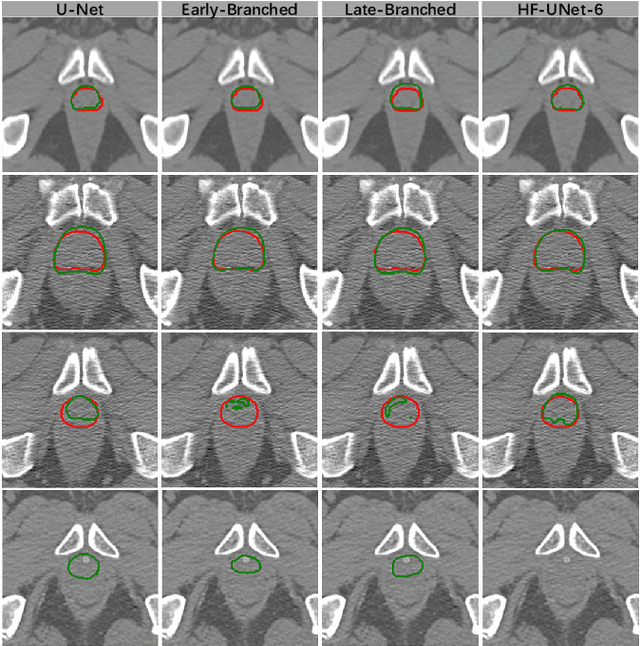
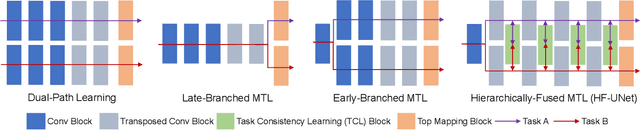
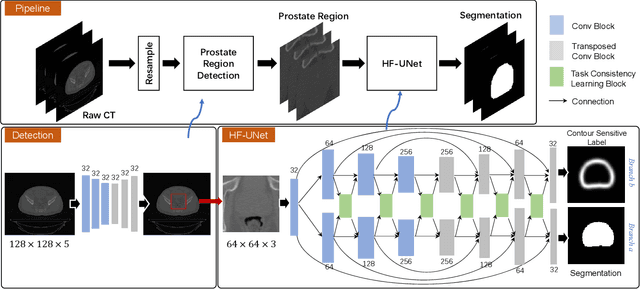
Abstract:Accurate segmentation of the prostate is a key step in external beam radiation therapy treatments. In this paper, we tackle the challenging task of prostate segmentation in CT images by a two-stage network with 1) the first stage to fast localize, and 2) the second stage to accurately segment the prostate. To precisely segment the prostate in the second stage, we formulate prostate segmentation into a multi-task learning framework, which includes a main task to segment the prostate, and an auxiliary task to delineate the prostate boundary. Here, the second task is applied to provide additional guidance of unclear prostate boundary in CT images. Besides, the conventional multi-task deep networks typically share most of the parameters (i.e., feature representations) across all tasks, which may limit their data fitting ability, as the specificities of different tasks are inevitably ignored. By contrast, we solve them by a hierarchically-fused U-Net structure, namely HF-UNet. The HF-UNet has two complementary branches for two tasks, with the novel proposed attention-based task consistency learning block to communicate at each level between the two decoding branches. Therefore, HF-UNet endows the ability to learn hierarchically the shared representations for different tasks, and preserve the specificities of learned representations for different tasks simultaneously. We did extensive evaluations of the proposed method on a large planning CT image dataset, including images acquired from 339 patients. The experimental results show HF-UNet outperforms the conventional multi-task network architectures and the state-of-the-art methods.
TripletUNet: Multi-Task U-Net with Online Voxel-Wise Learning for Precise CT Prostate Segmentation
May 21, 2020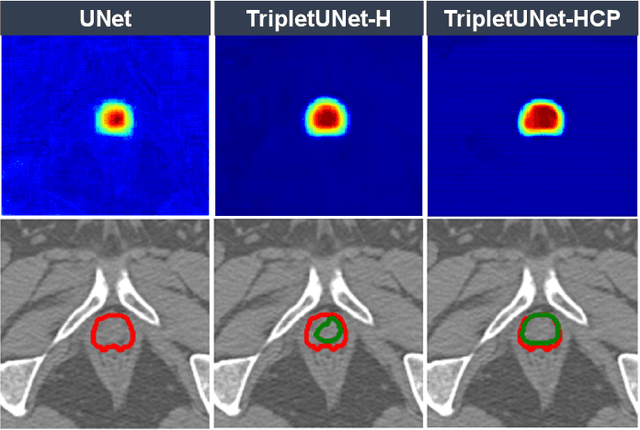
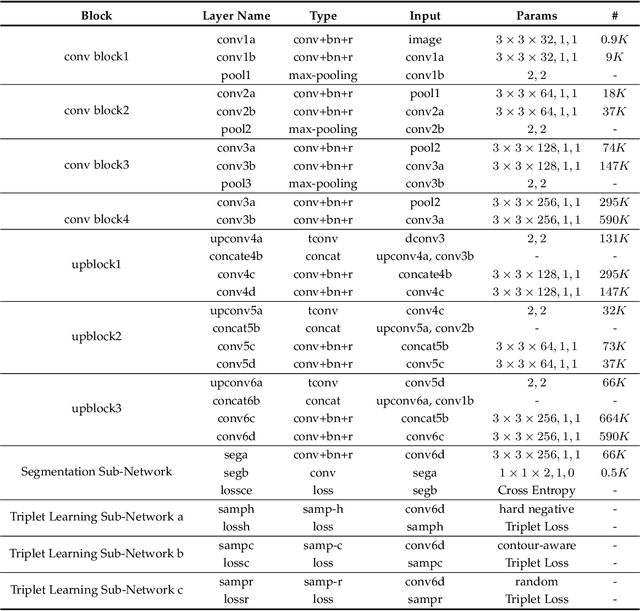
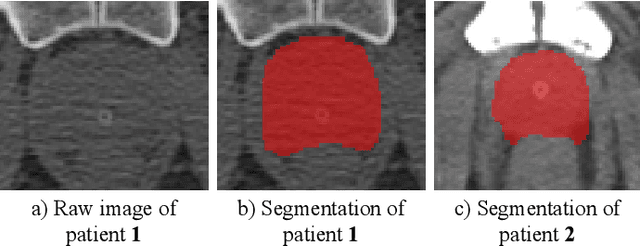
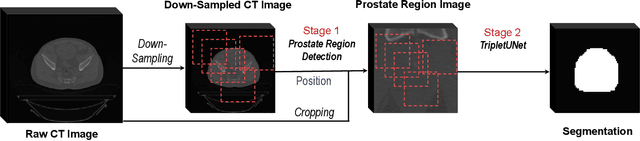
Abstract:Fully convolutional networks (FCNs), including U-Net and V-Net, are widely-used network architecture for semantic segmentation in recent studies. However, conventional FCNs are typically trained by the cross-entropy loss or dice loss, in which the relationships among voxels are neglected. This often results in non-smooth neighborhoods in the output segmentation map. This problem becomes more serious in CT prostate segmentation as CT images are usually of low tissue contrast. To address this problem, we propose a two-stage framework. The first stage quickly localizes the prostate region. Then, the second stage precisely segments the prostate by a multi-task FCN-based on the U-Net architecture. We introduce a novel online voxel-triplet learning module through metric learning and voxel feature embeddings in the multi-task network. The proposed network has two branches guided by two tasks: 1) a segmentation sub-network aiming to generate prostate segmentation, and 2) a triplet learning sub-network aiming to improve the quality of the learned feature space supervised by a mixed of triplet and pair-wise loss function. The triplet learning sub-network samples triplets from the inter-mediate heatmap. Unlike conventional deep triplet learning methods that generate triplets before the training phase, our proposed voxel-triplets are sampled in an online manner and operates in an end-to-end fashion via multi-task learning. To evaluate the proposed method, we implement comprehensive experiments on a CT image dataset consisting of 339 patients. The ablation studies show that our method can effectively learn more representative voxel-level features compared with the conventional FCN network. And the comparisons show that the proposed method outperforms the state-of-the-art methods by a large margin.
LEARN: Learned Experts' Assessment-based Reconstruction Network for Sparse-data CT
Feb 10, 2018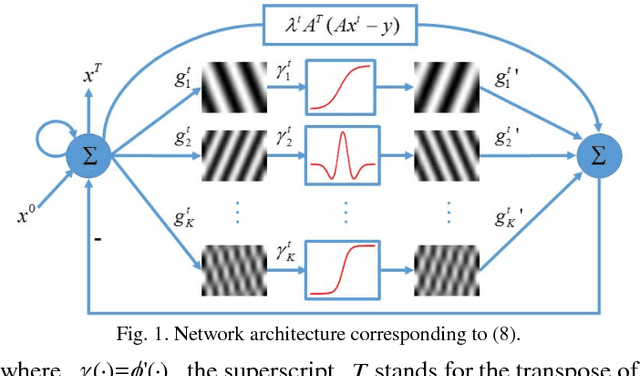
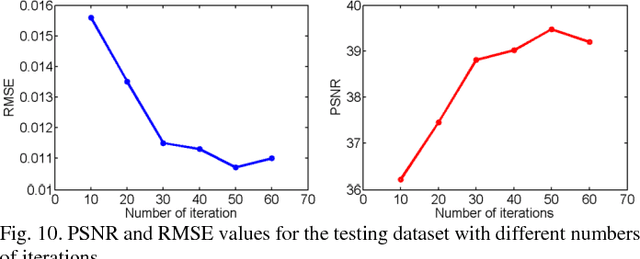
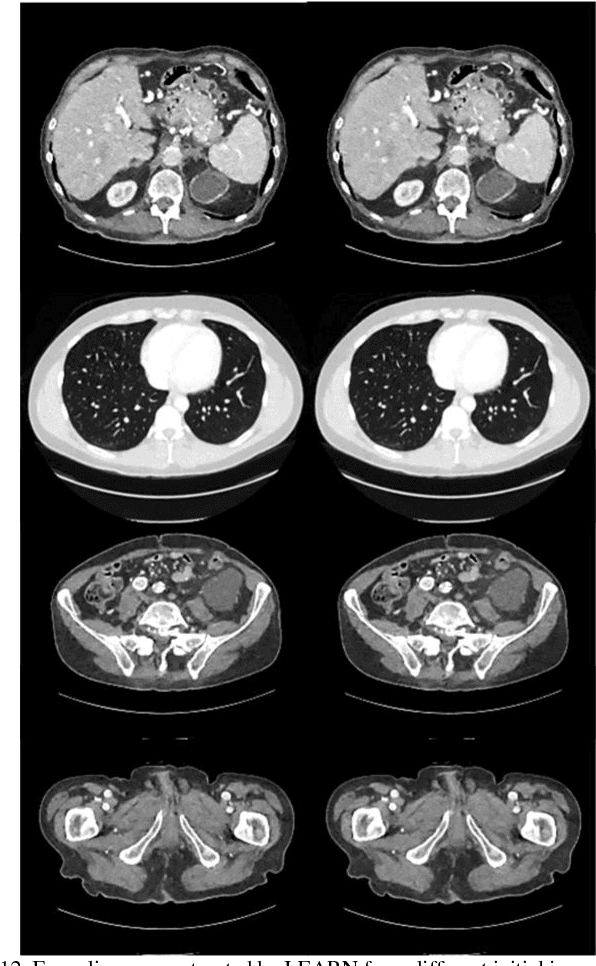
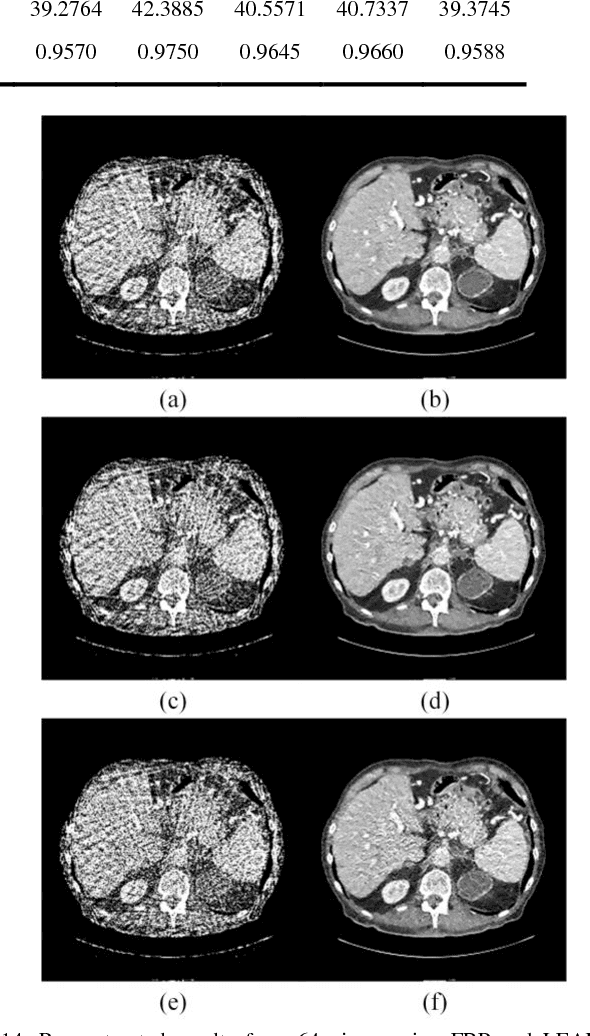
Abstract:Compressive sensing (CS) has proved effective for tomographic reconstruction from sparsely collected data or under-sampled measurements, which are practically important for few-view CT, tomosynthesis, interior tomography, and so on. To perform sparse-data CT, the iterative reconstruction commonly use regularizers in the CS framework. Currently, how to choose the parameters adaptively for regularization is a major open problem. In this paper, inspired by the idea of machine learning especially deep learning, we unfold a state-of-the-art "fields of experts" based iterative reconstruction scheme up to a number of iterations for data-driven training, construct a Learned Experts' Assessment-based Reconstruction Network ("LEARN") for sparse-data CT, and demonstrate the feasibility and merits of our LEARN network. The experimental results with our proposed LEARN network produces a competitive performance with the well-known Mayo Clinic Low-Dose Challenge Dataset relative to several state-of-the-art methods, in terms of artifact reduction, feature preservation, and computational speed. This is consistent to our insight that because all the regularization terms and parameters used in the iterative reconstruction are now learned from the training data, our LEARN network utilizes application-oriented knowledge more effectively and recovers underlying images more favorably than competing algorithms. Also, the number of layers in the LEARN network is only 12, reducing the computational complexity of typical iterative algorithms by orders of magnitude.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge