Xiaohuan Cao
SDF-Net: A Hybrid Detection Network for Mediastinal Lymph Node Detection on Contrast CT Images
Sep 10, 2024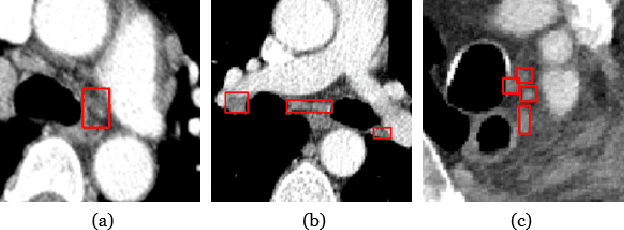
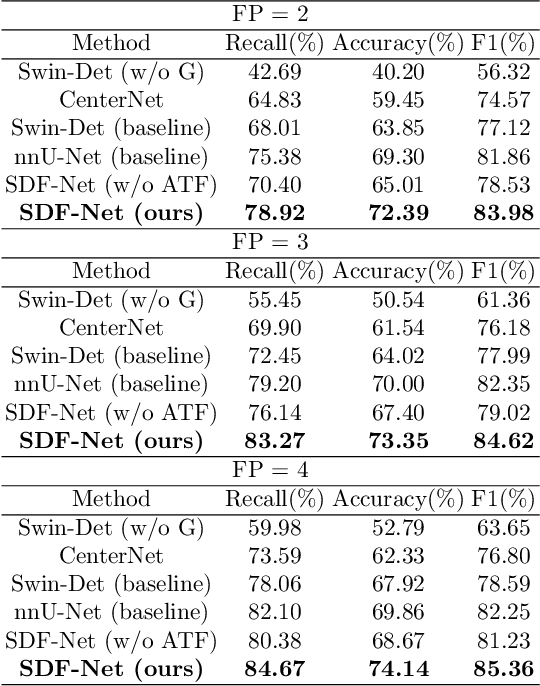
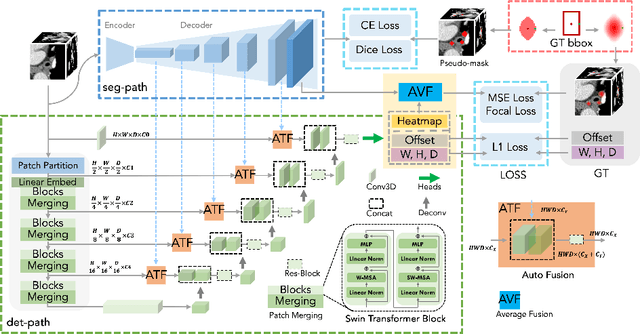
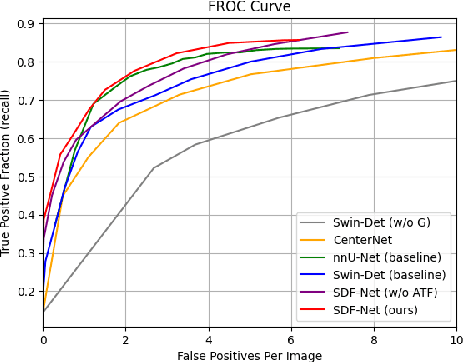
Abstract:Accurate lymph node detection and quantification are crucial for cancer diagnosis and staging on contrast-enhanced CT images, as they impact treatment planning and prognosis. However, detecting lymph nodes in the mediastinal area poses challenges due to their low contrast, irregular shapes and dispersed distribution. In this paper, we propose a Swin-Det Fusion Network (SDF-Net) to effectively detect lymph nodes. SDF-Net integrates features from both segmentation and detection to enhance the detection capability of lymph nodes with various shapes and sizes. Specifically, an auto-fusion module is designed to merge the feature maps of segmentation and detection networks at different levels. To facilitate effective learning without mask annotations, we introduce a shape-adaptive Gaussian kernel to represent lymph node in the training stage and provide more anatomical information for effective learning. Comparative results demonstrate promising performance in addressing the complex lymph node detection problem.
Structure-aware registration network for liver DCE-CT images
Mar 08, 2023



Abstract:Image registration of liver dynamic contrast-enhanced computed tomography (DCE-CT) is crucial for diagnosis and image-guided surgical planning of liver cancer. However, intensity variations due to the flow of contrast agents combined with complex spatial motion induced by respiration brings great challenge to existing intensity-based registration methods. To address these problems, we propose a novel structure-aware registration method by incorporating structural information of related organs with segmentation-guided deep registration network. Existing segmentation-guided registration methods only focus on volumetric registration inside the paired organ segmentations, ignoring the inherent attributes of their anatomical structures. In addition, such paired organ segmentations are not always available in DCE-CT images due to the flow of contrast agents. Different from existing segmentation-guided registration methods, our proposed method extracts structural information in hierarchical geometric perspectives of line and surface. Then, according to the extracted structural information, structure-aware constraints are constructed and imposed on the forward and backward deformation field simultaneously. In this way, all available organ segmentations, including unpaired ones, can be fully utilized to avoid the side effect of contrast agent and preserve the topology of organs during registration. Extensive experiments on an in-house liver DCE-CT dataset and a public LiTS dataset show that our proposed method can achieve higher registration accuracy and preserve anatomical structure more effectively than state-of-the-art methods.
Sentiment Word Aware Multimodal Refinement for Multimodal Sentiment Analysis with ASR Errors
Mar 01, 2022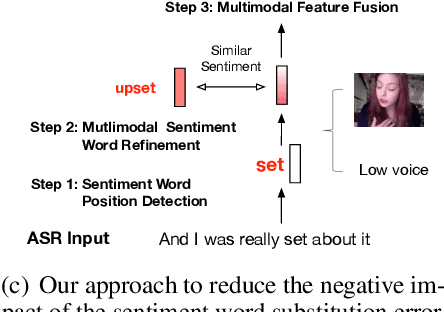
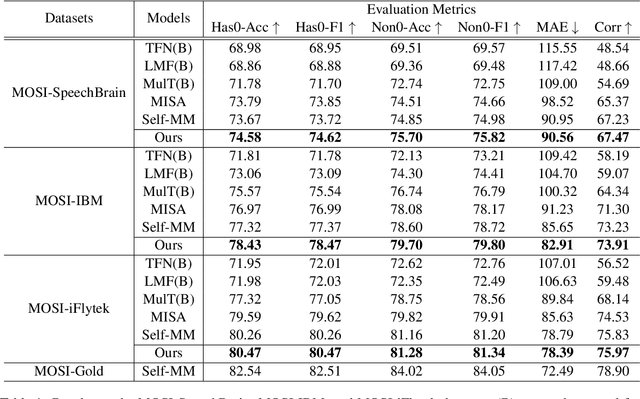
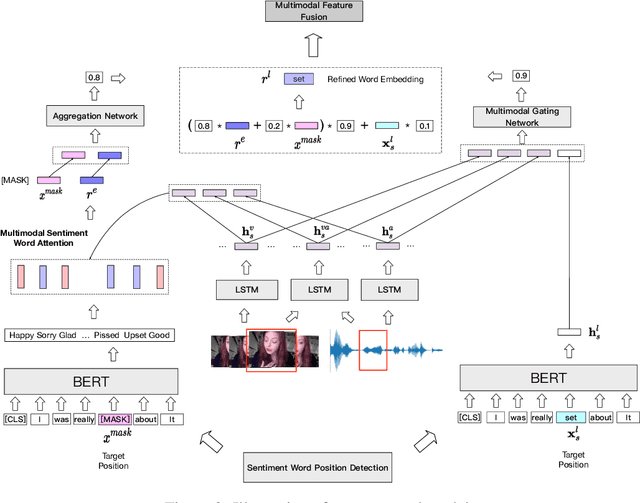
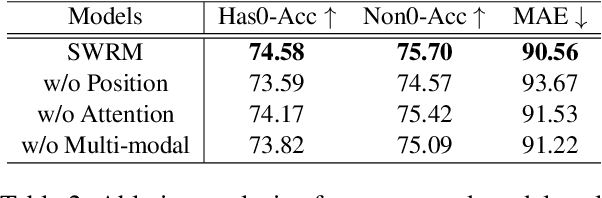
Abstract:Multimodal sentiment analysis has attracted increasing attention and lots of models have been proposed. However, the performance of the state-of-the-art models decreases sharply when they are deployed in the real world. We find that the main reason is that real-world applications can only access the text outputs by the automatic speech recognition (ASR) models, which may be with errors because of the limitation of model capacity. Through further analysis of the ASR outputs, we find that in some cases the sentiment words, the key sentiment elements in the textual modality, are recognized as other words, which makes the sentiment of the text change and hurts the performance of multimodal sentiment models directly. To address this problem, we propose the sentiment word aware multimodal refinement model (SWRM), which can dynamically refine the erroneous sentiment words by leveraging multimodal sentiment clues. Specifically, we first use the sentiment word position detection module to obtain the most possible position of the sentiment word in the text and then utilize the multimodal sentiment word refinement module to dynamically refine the sentiment word embeddings. The refined embeddings are taken as the textual inputs of the multimodal feature fusion module to predict the sentiment labels. We conduct extensive experiments on the real-world datasets including MOSI-Speechbrain, MOSI-IBM, and MOSI-iFlytek and the results demonstrate the effectiveness of our model, which surpasses the current state-of-the-art models on three datasets. Furthermore, our approach can be adapted for other multimodal feature fusion models easily. Data and code are available at https://github.com/albertwy/SWRM.
HF-UNet: Learning Hierarchically Inter-Task Relevance in Multi-Task U-Net for Accurate Prostate Segmentation
May 23, 2020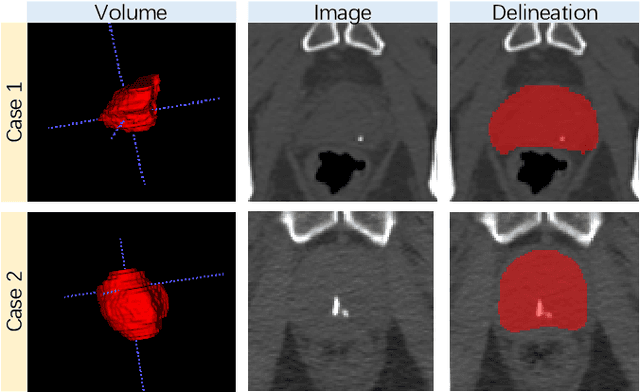
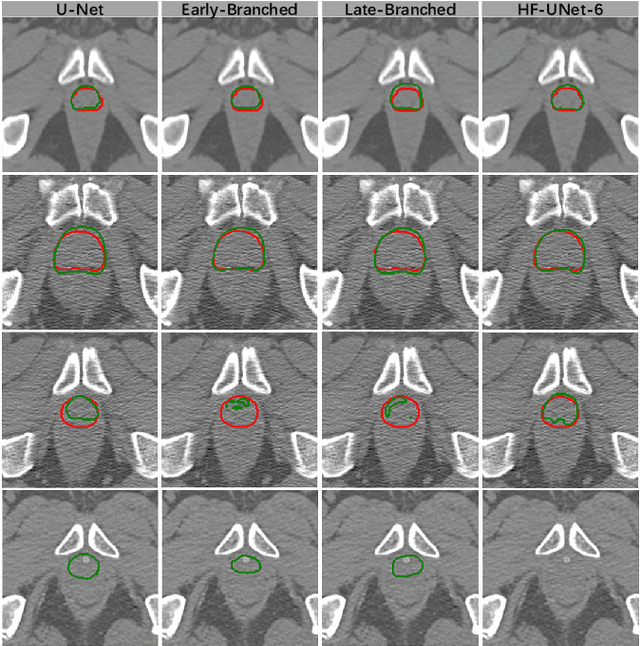
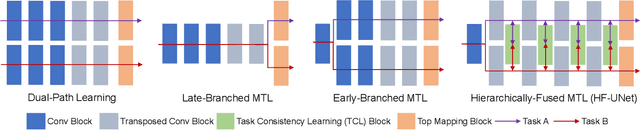
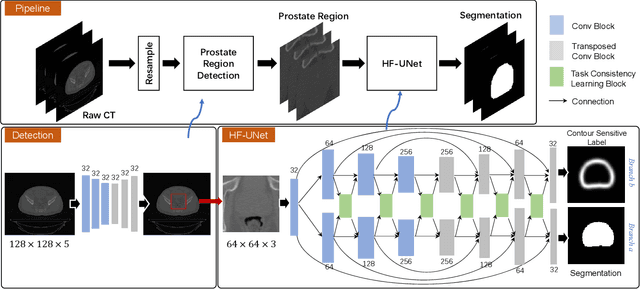
Abstract:Accurate segmentation of the prostate is a key step in external beam radiation therapy treatments. In this paper, we tackle the challenging task of prostate segmentation in CT images by a two-stage network with 1) the first stage to fast localize, and 2) the second stage to accurately segment the prostate. To precisely segment the prostate in the second stage, we formulate prostate segmentation into a multi-task learning framework, which includes a main task to segment the prostate, and an auxiliary task to delineate the prostate boundary. Here, the second task is applied to provide additional guidance of unclear prostate boundary in CT images. Besides, the conventional multi-task deep networks typically share most of the parameters (i.e., feature representations) across all tasks, which may limit their data fitting ability, as the specificities of different tasks are inevitably ignored. By contrast, we solve them by a hierarchically-fused U-Net structure, namely HF-UNet. The HF-UNet has two complementary branches for two tasks, with the novel proposed attention-based task consistency learning block to communicate at each level between the two decoding branches. Therefore, HF-UNet endows the ability to learn hierarchically the shared representations for different tasks, and preserve the specificities of learned representations for different tasks simultaneously. We did extensive evaluations of the proposed method on a large planning CT image dataset, including images acquired from 339 patients. The experimental results show HF-UNet outperforms the conventional multi-task network architectures and the state-of-the-art methods.
Dual-Sampling Attention Network for Diagnosis of COVID-19 from Community Acquired Pneumonia
May 20, 2020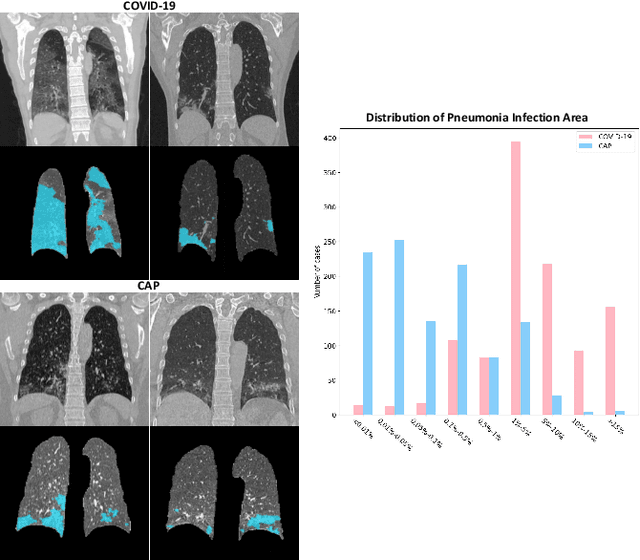
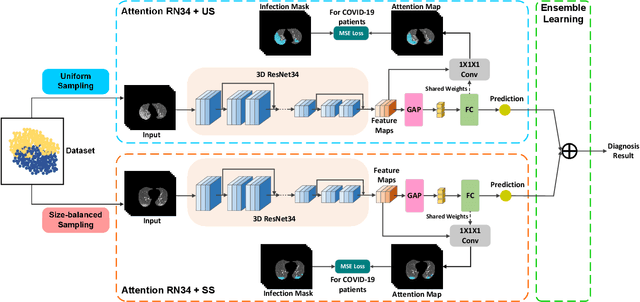
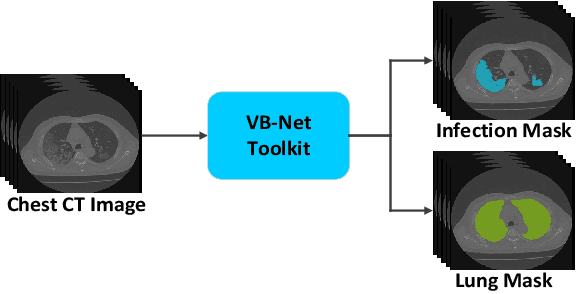
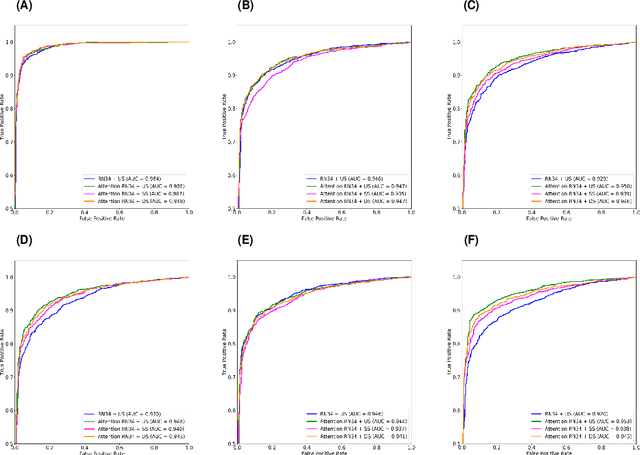
Abstract:The coronavirus disease (COVID-19) is rapidly spreading all over the world, and has infected more than 1,436,000 people in more than 200 countries and territories as of April 9, 2020. Detecting COVID-19 at early stage is essential to deliver proper healthcare to the patients and also to protect the uninfected population. To this end, we develop a dual-sampling attention network to automatically diagnose COVID- 19 from the community acquired pneumonia (CAP) in chest computed tomography (CT). In particular, we propose a novel online attention module with a 3D convolutional network (CNN) to focus on the infection regions in lungs when making decisions of diagnoses. Note that there exists imbalanced distribution of the sizes of the infection regions between COVID-19 and CAP, partially due to fast progress of COVID-19 after symptom onset. Therefore, we develop a dual-sampling strategy to mitigate the imbalanced learning. Our method is evaluated (to our best knowledge) upon the largest multi-center CT data for COVID-19 from 8 hospitals. In the training-validation stage, we collect 2186 CT scans from 1588 patients for a 5-fold cross-validation. In the testing stage, we employ another independent large-scale testing dataset including 2796 CT scans from 2057 patients. Results show that our algorithm can identify the COVID-19 images with the area under the receiver operating characteristic curve (AUC) value of 0.944, accuracy of 87.5%, sensitivity of 86.9%, specificity of 90.1%, and F1-score of 82.0%. With this performance, the proposed algorithm could potentially aid radiologists with COVID-19 diagnosis from CAP, especially in the early stage of the COVID-19 outbreak.
Deep Morphological Simplification Network (MS-Net) for Guided Registration of Brain Magnetic Resonance Images
Feb 06, 2019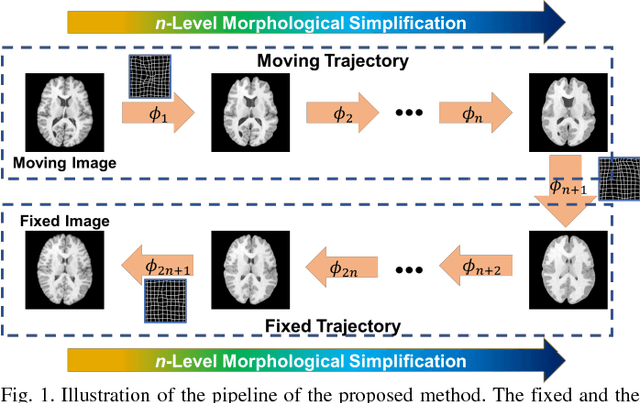
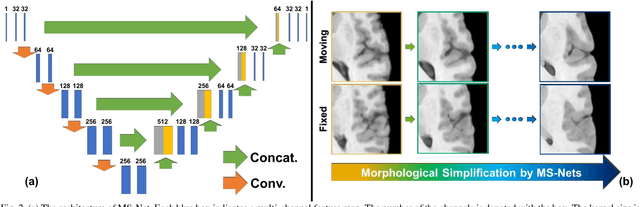
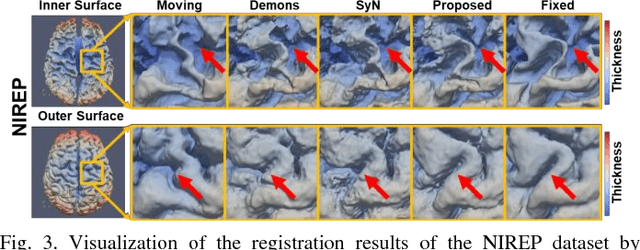
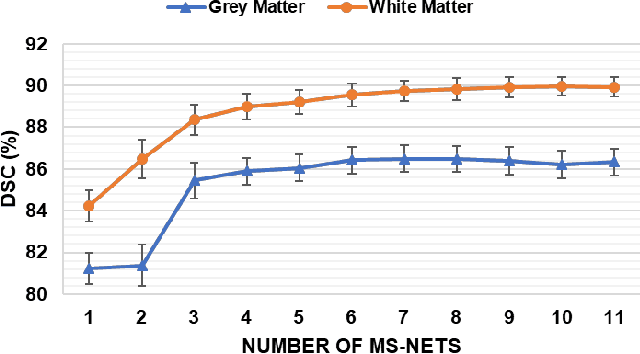
Abstract:Objective: Deformable brain MR image registration is challenging due to large inter-subject anatomical variation. For example, the highly complex cortical folding pattern makes it hard to accurately align corresponding cortical structures of individual images. In this paper, we propose a novel deep learning way to simplify the difficult registration problem of brain MR images. Methods: We train a morphological simplification network (MS-Net), which can generate a "simple" image with less anatomical details based on the "complex" input. With MS-Net, the complexity of the fixed image or the moving image under registration can be reduced gradually, thus building an individual (simplification) trajectory represented by MS-Net outputs. Since the generated images at the ends of the two trajectories (of the fixed and moving images) are so simple and very similar in appearance, they are easy to register. Thus, the two trajectories can act as a bridge to link the fixed and the moving images, and guide their registration. Results: Our experiments show that the proposed method can achieve highly accurate registration performance on different datasets (i.e., NIREP, LPBA, IBSR, CUMC, and MGH). Moreover, the method can be also easily transferred across diverse image datasets and obtain superior accuracy on surface alignment. Conclusion and Significance: We propose MS-Net as a powerful and flexible tool to simplify brain MR images and their registration. To our knowledge, this is the first work to simplify brain MR image registration by deep learning, instead of estimating deformation field directly.
Deep Learning based Inter-Modality Image Registration Supervised by Intra-Modality Similarity
Apr 28, 2018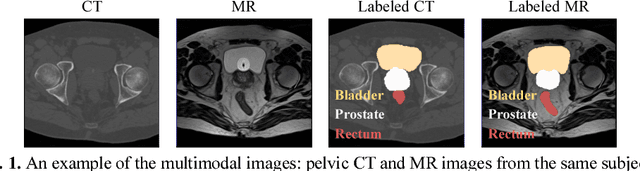
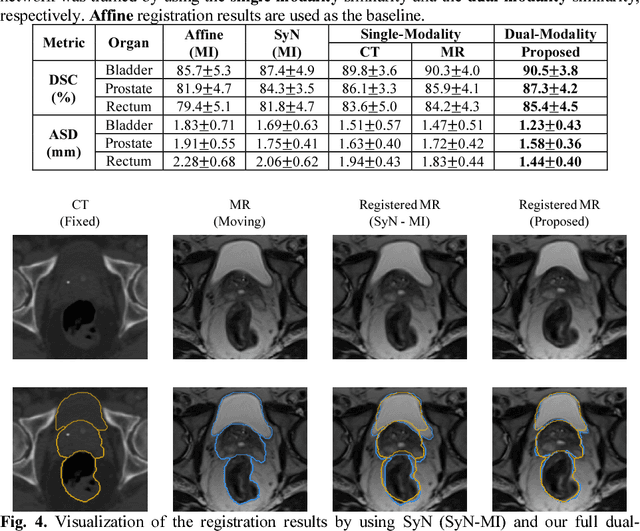
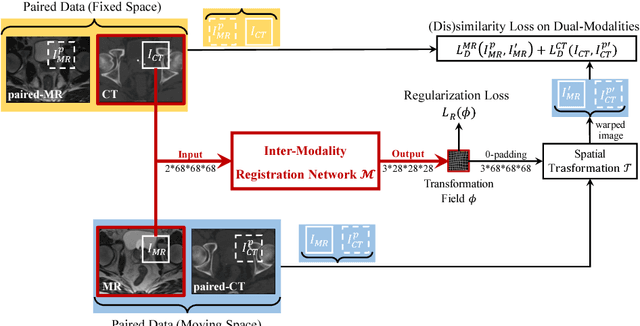
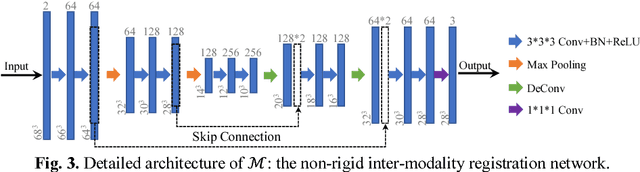
Abstract:Non-rigid inter-modality registration can facilitate accurate information fusion from different modalities, but it is challenging due to the very different image appearances across modalities. In this paper, we propose to train a non-rigid inter-modality image registration network, which can directly predict the transformation field from the input multimodal images, such as CT and MR images. In particular, the training of our inter-modality registration network is supervised by intra-modality similarity metric based on the available paired data, which is derived from a pre-aligned CT and MR dataset. Specifically, in the training stage, to register the input CT and MR images, their similarity is evaluated on the warped MR image and the MR image that is paired with the input CT. So that, the intra-modality similarity metric can be directly applied to measure whether the input CT and MR images are well registered. Moreover, we use the idea of dual-modality fashion, in which we measure the similarity on both CT modality and MR modality. In this way, the complementary anatomies in both modalities can be jointly considered to more accurately train the inter-modality registration network. In the testing stage, the trained inter-modality registration network can be directly applied to register the new multimodal images without any paired data. Experimental results have shown that, the proposed method can achieve promising accuracy and efficiency for the challenging non-rigid inter-modality registration task and also outperforms the state-of-the-art approaches.
BIRNet: Brain Image Registration Using Dual-Supervised Fully Convolutional Networks
Feb 13, 2018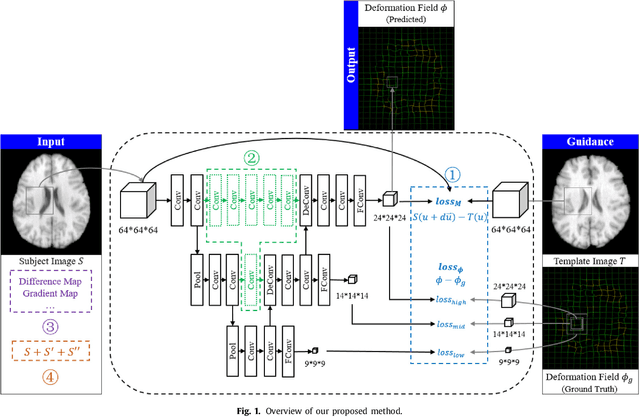

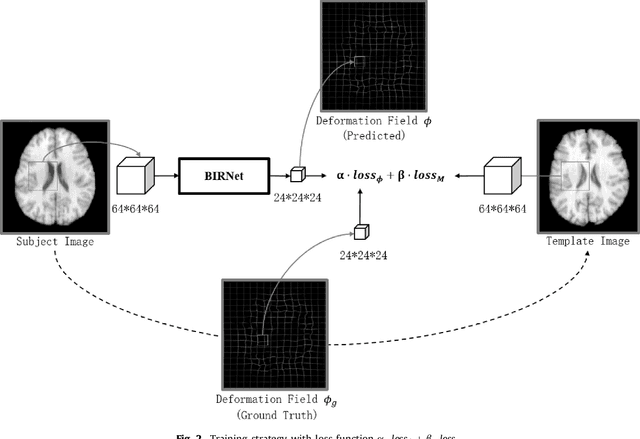
Abstract:In this paper, we propose a deep learning approach for image registration by predicting deformation from image appearance. Since obtaining ground-truth deformation fields for training can be challenging, we design a fully convolutional network that is subject to dual-guidance: (1) Coarse guidance using deformation fields obtained by an existing registration method; and (2) Fine guidance using image similarity. The latter guidance helps avoid overly relying on the supervision from the training deformation fields, which could be inaccurate. For effective training, we further improve the deep convolutional network with gap filling, hierarchical loss, and multi-source strategies. Experiments on a variety of datasets show promising registration accuracy and efficiency compared with state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge