Dongming Wei
Unsupervised Landmark Detection Based Spatiotemporal Motion Estimation for 4D Dynamic Medical Images
Oct 12, 2021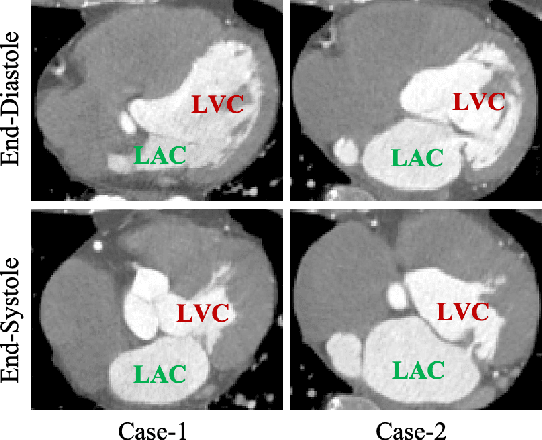
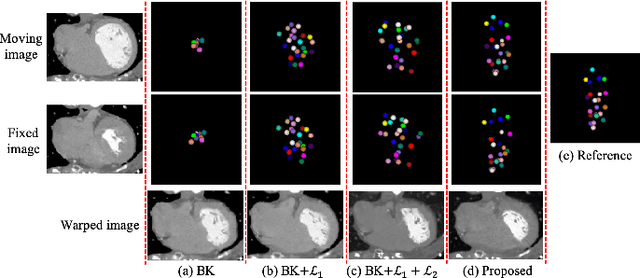
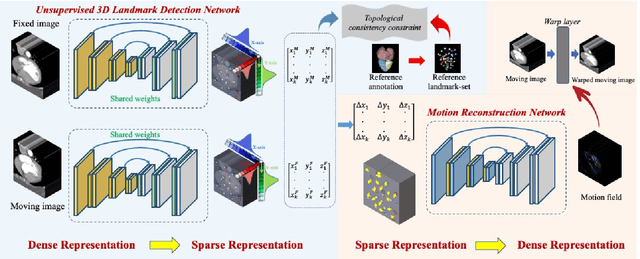
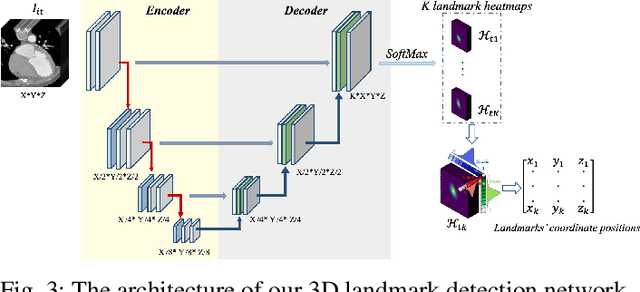
Abstract:Motion estimation is a fundamental step in dynamic medical image processing for the assessment of target organ anatomy and function. However, existing image-based motion estimation methods, which optimize the motion field by evaluating the local image similarity, are prone to produce implausible estimation, especially in the presence of large motion. In this study, we provide a novel motion estimation framework of Dense-Sparse-Dense (DSD), which comprises two stages. In the first stage, we process the raw dense image to extract sparse landmarks to represent the target organ anatomical topology and discard the redundant information that is unnecessary for motion estimation. For this purpose, we introduce an unsupervised 3D landmark detection network to extract spatially sparse but representative landmarks for the target organ motion estimation. In the second stage, we derive the sparse motion displacement from the extracted sparse landmarks of two images of different time points. Then, we present a motion reconstruction network to construct the motion field by projecting the sparse landmarks displacement back into the dense image domain. Furthermore, we employ the estimated motion field from our two-stage DSD framework as initialization and boost the motion estimation quality in light-weight yet effective iterative optimization. We evaluate our method on two dynamic medical imaging tasks to model cardiac motion and lung respiratory motion, respectively. Our method has produced superior motion estimation accuracy compared to existing comparative methods. Besides, the extensive experimental results demonstrate that our solution can extract well representative anatomical landmarks without any requirement of manual annotation. Our code is publicly available online.
An Auto-Context Deformable Registration Network for Infant Brain MRI
May 19, 2020



Abstract:Deformable image registration is fundamental to longitudinal and population analysis. Geometric alignment of the infant brain MR images is challenging, owing to rapid changes in image appearance in association with brain development. In this paper, we propose an infant-dedicated deep registration network that uses the auto-context strategy to gradually refine the deformation fields to obtain highly accurate correspondences. Instead of training multiple registration networks, our method estimates the deformation fields by invoking a single network multiple times for iterative deformation refinement. The final deformation field is obtained by the incremental composition of the deformation fields. Experimental results in comparison with state-of-the-art registration methods indicate that our method achieves higher accuracy while at the same time preserves the smoothness of the deformation fields. Our implementation is available online.
Robust Brain Magnetic Resonance Image Segmentation for Hydrocephalus Patients: Hard and Soft Attention
Jan 12, 2020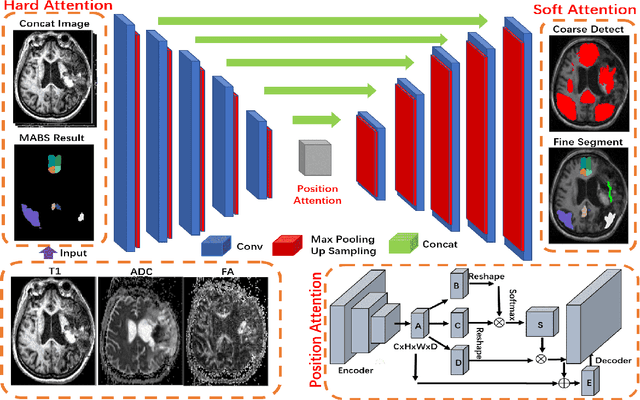
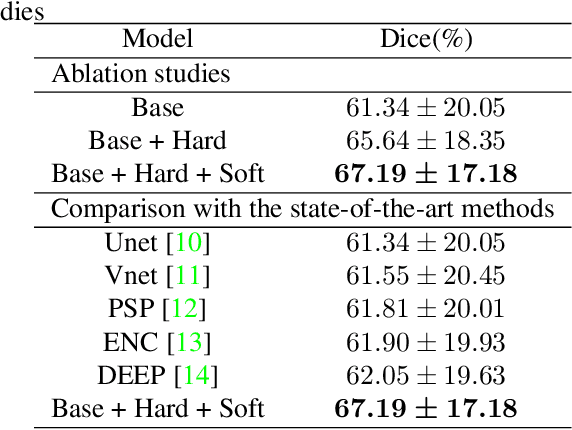
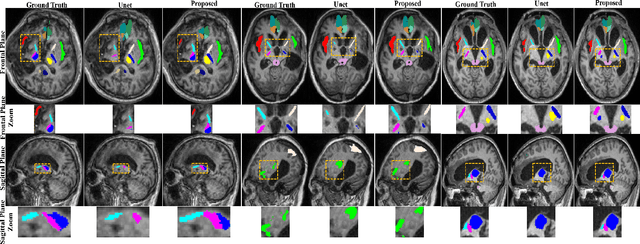
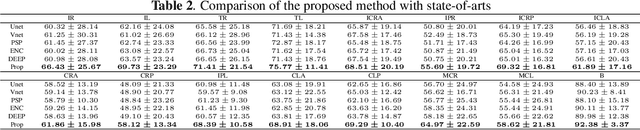
Abstract:Brain magnetic resonance (MR) segmentation for hydrocephalus patients is considered as a challenging work. Encoding the variation of the brain anatomical structures from different individuals cannot be easily achieved. The task becomes even more difficult especially when the image data from hydrocephalus patients are considered, which often have large deformations and differ significantly from the normal subjects. Here, we propose a novel strategy with hard and soft attention modules to solve the segmentation problems for hydrocephalus MR images. Our main contributions are three-fold: 1) the hard-attention module generates coarse segmentation map using multi-atlas-based method and the VoxelMorph tool, which guides subsequent segmentation process and improves its robustness; 2) the soft-attention module incorporates position attention to capture precise context information, which further improves the segmentation accuracy; 3) we validate our method by segmenting insula, thalamus and many other regions-of-interests (ROIs) that are critical to quantify brain MR images of hydrocephalus patients in real clinical scenario. The proposed method achieves much improved robustness and accuracy when segmenting all 17 consciousness-related ROIs with high variations for different subjects. To the best of our knowledge, this is the first work to employ deep learning for solving the brain segmentation problems of hydrocephalus patients.
Synthesis and Inpainting-Based MR-CT Registration for Image-Guided Thermal Ablation of Liver Tumors
Jul 30, 2019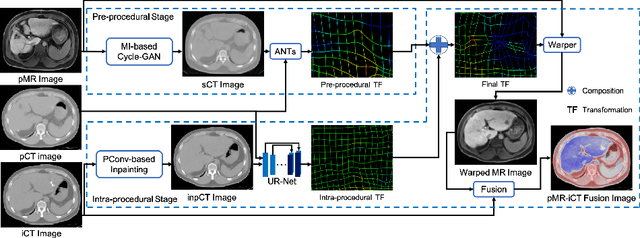
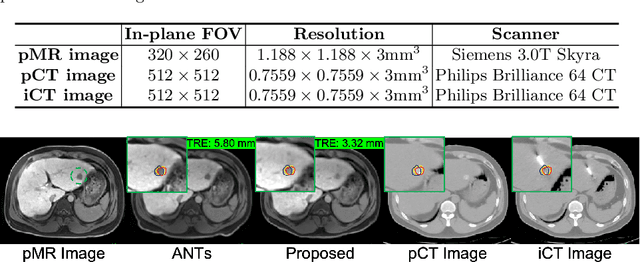
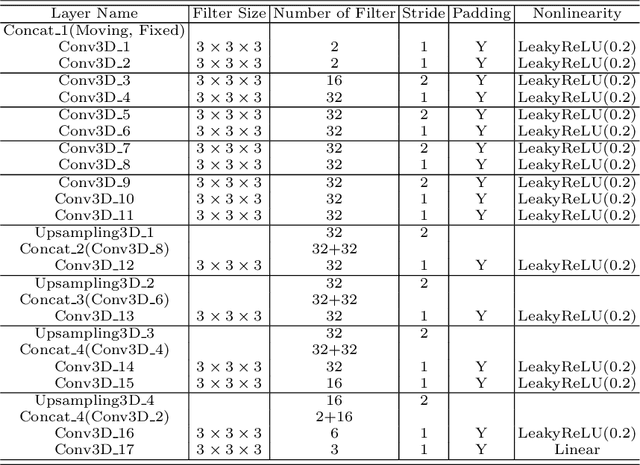
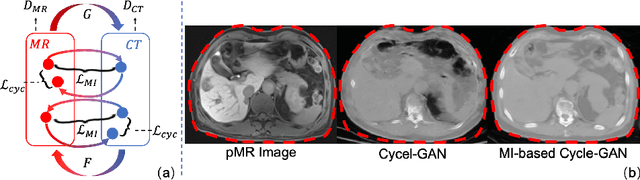
Abstract:Thermal ablation is a minimally invasive procedure for treat-ing small or unresectable tumors. Although CT is widely used for guiding ablation procedures, the contrast of tumors against surrounding normal tissues in CT images is often poor, aggravating the difficulty in accurate thermal ablation. In this paper, we propose a fast MR-CT image registration method to overlay a pre-procedural MR (pMR) image onto an intra-procedural CT (iCT) image for guiding the thermal ablation of liver tumors. By first using a Cycle-GAN model with mutual information constraint to generate synthesized CT (sCT) image from the cor-responding pMR, pre-procedural MR-CT image registration is carried out through traditional mono-modality CT-CT image registration. At the intra-procedural stage, a partial-convolution-based network is first used to inpaint the probe and its artifacts in the iCT image. Then, an unsupervised registration network is used to efficiently align the pre-procedural CT (pCT) with the inpainted iCT (inpCT) image. The final transformation from pMR to iCT is obtained by combining the two estimated transformations,i.e., (1) from the pMR image space to the pCT image space (through sCT) and (2) from the pCT image space to the iCT image space (through inpCT). Experimental results confirm that the proposed method achieves high registration accuracy with a very fast computational speed.
Deep Morphological Simplification Network (MS-Net) for Guided Registration of Brain Magnetic Resonance Images
Feb 06, 2019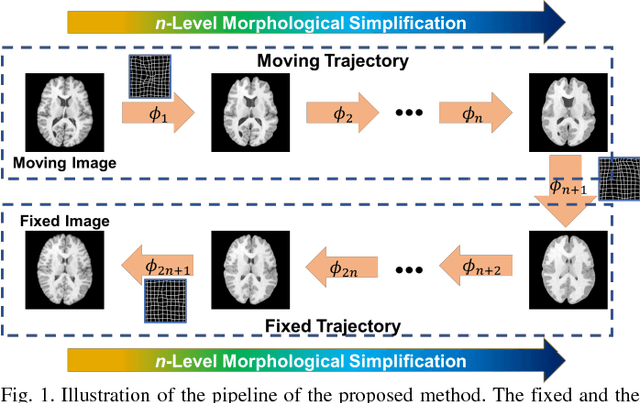
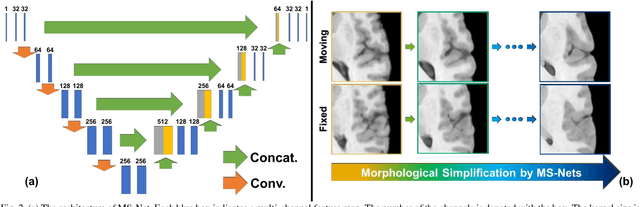
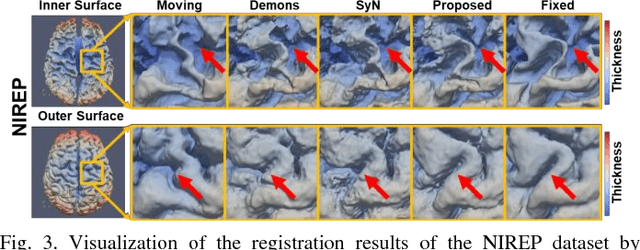
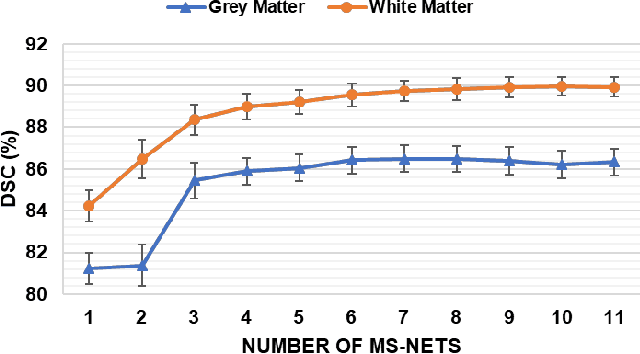
Abstract:Objective: Deformable brain MR image registration is challenging due to large inter-subject anatomical variation. For example, the highly complex cortical folding pattern makes it hard to accurately align corresponding cortical structures of individual images. In this paper, we propose a novel deep learning way to simplify the difficult registration problem of brain MR images. Methods: We train a morphological simplification network (MS-Net), which can generate a "simple" image with less anatomical details based on the "complex" input. With MS-Net, the complexity of the fixed image or the moving image under registration can be reduced gradually, thus building an individual (simplification) trajectory represented by MS-Net outputs. Since the generated images at the ends of the two trajectories (of the fixed and moving images) are so simple and very similar in appearance, they are easy to register. Thus, the two trajectories can act as a bridge to link the fixed and the moving images, and guide their registration. Results: Our experiments show that the proposed method can achieve highly accurate registration performance on different datasets (i.e., NIREP, LPBA, IBSR, CUMC, and MGH). Moreover, the method can be also easily transferred across diverse image datasets and obtain superior accuracy on surface alignment. Conclusion and Significance: We propose MS-Net as a powerful and flexible tool to simplify brain MR images and their registration. To our knowledge, this is the first work to simplify brain MR image registration by deep learning, instead of estimating deformation field directly.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge