Dagan Feng
MedCFVQA: A Causal Approach to Mitigate Modality Preference Bias in Medical Visual Question Answering
May 23, 2025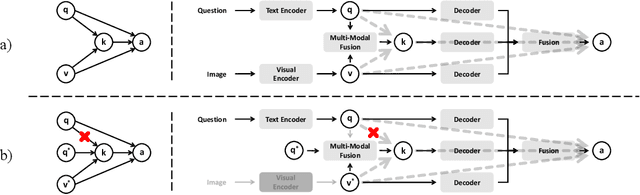
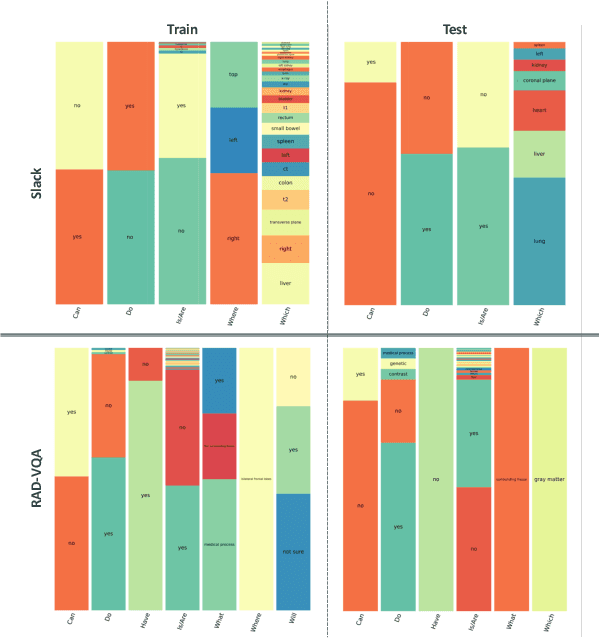
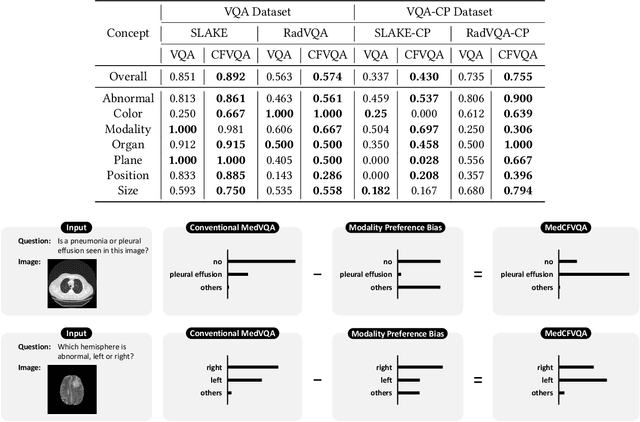
Abstract:Medical Visual Question Answering (MedVQA) is crucial for enhancing the efficiency of clinical diagnosis by providing accurate and timely responses to clinicians' inquiries regarding medical images. Existing MedVQA models suffered from modality preference bias, where predictions are heavily dominated by one modality while overlooking the other (in MedVQA, usually questions dominate the answer but images are overlooked), thereby failing to learn multimodal knowledge. To overcome the modality preference bias, we proposed a Medical CounterFactual VQA (MedCFVQA) model, which trains with bias and leverages causal graphs to eliminate the modality preference bias during inference. Existing MedVQA datasets exhibit substantial prior dependencies between questions and answers, which results in acceptable performance even if the model significantly suffers from the modality preference bias. To address this issue, we reconstructed new datasets by leveraging existing MedVQA datasets and Changed their P3rior dependencies (CP) between questions and their answers in the training and test set. Extensive experiments demonstrate that MedCFVQA significantly outperforms its non-causal counterpart on both SLAKE, RadVQA and SLAKE-CP, RadVQA-CP datasets.
A Causal Approach to Mitigate Modality Preference Bias in Medical Visual Question Answering
May 22, 2025


Abstract:Medical Visual Question Answering (MedVQA) is crucial for enhancing the efficiency of clinical diagnosis by providing accurate and timely responses to clinicians' inquiries regarding medical images. Existing MedVQA models suffered from modality preference bias, where predictions are heavily dominated by one modality while overlooking the other (in MedVQA, usually questions dominate the answer but images are overlooked), thereby failing to learn multimodal knowledge. To overcome the modality preference bias, we proposed a Medical CounterFactual VQA (MedCFVQA) model, which trains with bias and leverages causal graphs to eliminate the modality preference bias during inference. Existing MedVQA datasets exhibit substantial prior dependencies between questions and answers, which results in acceptable performance even if the model significantly suffers from the modality preference bias. To address this issue, we reconstructed new datasets by leveraging existing MedVQA datasets and Changed their P3rior dependencies (CP) between questions and their answers in the training and test set. Extensive experiments demonstrate that MedCFVQA significantly outperforms its non-causal counterpart on both SLAKE, RadVQA and SLAKE-CP, RadVQA-CP datasets.
3DPX: Single Panoramic X-ray Analysis Guided by 3D Oral Structure Reconstruction
Sep 27, 2024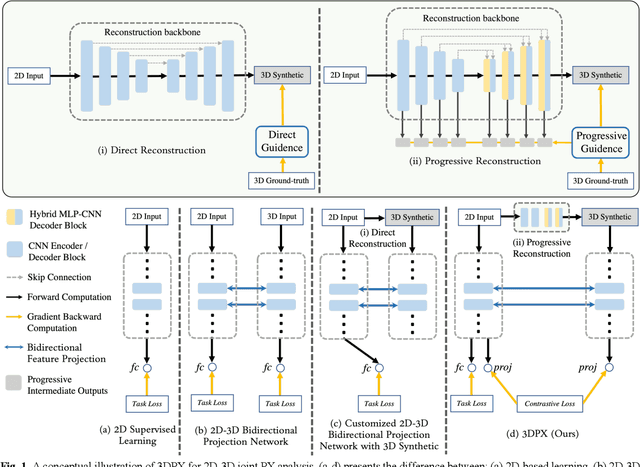
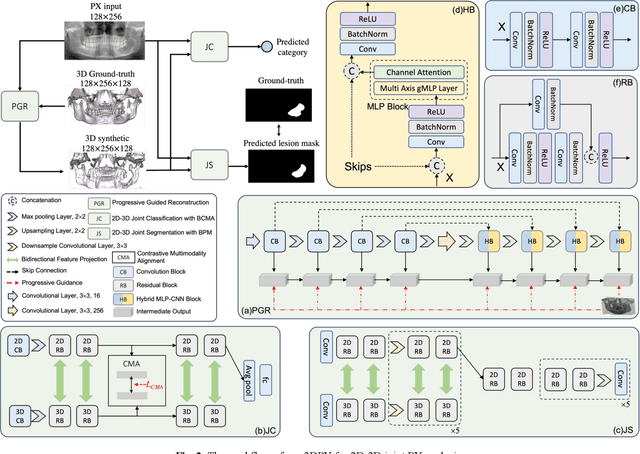
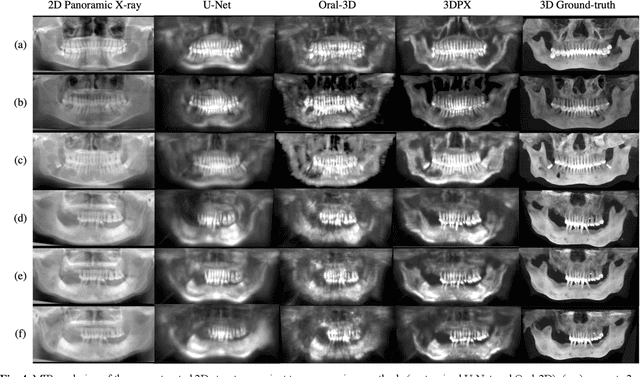
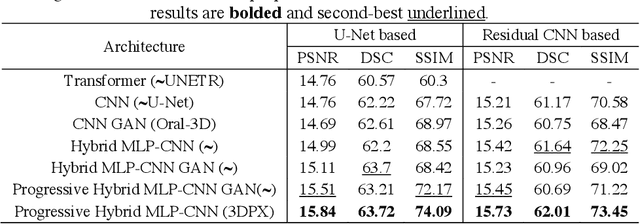
Abstract:Panoramic X-ray (PX) is a prevalent modality in dentistry practice owing to its wide availability and low cost. However, as a 2D projection of a 3D structure, PX suffers from anatomical information loss and PX diagnosis is limited compared to that with 3D imaging modalities. 2D-to-3D reconstruction methods have been explored for the ability to synthesize the absent 3D anatomical information from 2D PX for use in PX image analysis. However, there are challenges in leveraging such 3D synthesized reconstructions. First, inferring 3D depth from 2D images remains a challenging task with limited accuracy. The second challenge is the joint analysis of 2D PX with its 3D synthesized counterpart, with the aim to maximize the 2D-3D synergy while minimizing the errors arising from the synthesized image. In this study, we propose a new method termed 3DPX - PX image analysis guided by 2D-to-3D reconstruction, to overcome these challenges. 3DPX consists of (i) a novel progressive reconstruction network to improve 2D-to-3D reconstruction and, (ii) a contrastive-guided bidirectional multimodality alignment module for 3D-guided 2D PX classification and segmentation tasks. The reconstruction network progressively reconstructs 3D images with knowledge imposed on the intermediate reconstructions at multiple pyramid levels and incorporates Multilayer Perceptrons to improve semantic understanding. The downstream networks leverage the reconstructed images as 3D anatomical guidance to the PX analysis through feature alignment, which increases the 2D-3D synergy with bidirectional feature projection and decease the impact of potential errors with contrastive guidance. Extensive experiments on two oral datasets involving 464 studies demonstrate that 3DPX outperforms the state-of-the-art methods in various tasks including 2D-to-3D reconstruction, PX classification and lesion segmentation.
SGSeg: Enabling Text-free Inference in Language-guided Segmentation of Chest X-rays via Self-guidance
Sep 07, 2024Abstract:Segmentation of infected areas in chest X-rays is pivotal for facilitating the accurate delineation of pulmonary structures and pathological anomalies. Recently, multi-modal language-guided image segmentation methods have emerged as a promising solution for chest X-rays where the clinical text reports, depicting the assessment of the images, are used as guidance. Nevertheless, existing language-guided methods require clinical reports alongside the images, and hence, they are not applicable for use in image segmentation in a decision support context, but rather limited to retrospective image analysis after clinical reporting has been completed. In this study, we propose a self-guided segmentation framework (SGSeg) that leverages language guidance for training (multi-modal) while enabling text-free inference (uni-modal), which is the first that enables text-free inference in language-guided segmentation. We exploit the critical location information of both pulmonary and pathological structures depicted in the text reports and introduce a novel localization-enhanced report generation (LERG) module to generate clinical reports for self-guidance. Our LERG integrates an object detector and a location-based attention aggregator, weakly-supervised by a location-aware pseudo-label extraction module. Extensive experiments on a well-benchmarked QaTa-COV19 dataset demonstrate that our SGSeg achieved superior performance than existing uni-modal segmentation methods and closely matched the state-of-the-art performance of multi-modal language-guided segmentation methods.
3DPX: Progressive 2D-to-3D Oral Image Reconstruction with Hybrid MLP-CNN Networks
Aug 02, 2024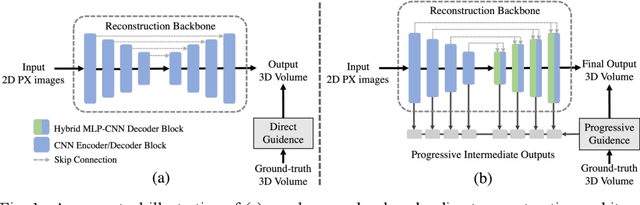
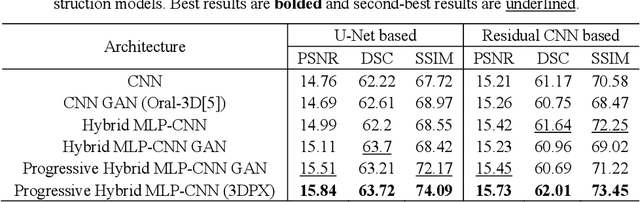
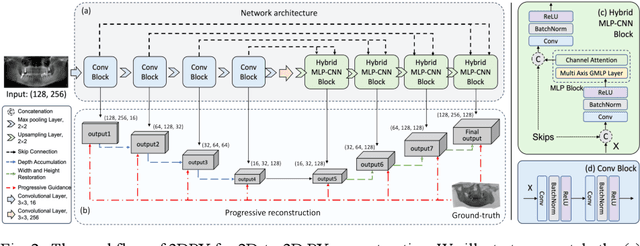
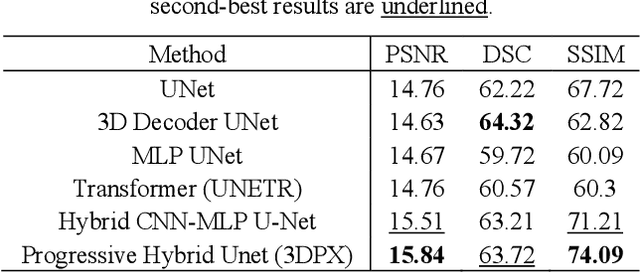
Abstract:Panoramic X-ray (PX) is a prevalent modality in dental practice for its wide availability and low cost. However, as a 2D projection image, PX does not contain 3D anatomical information, and therefore has limited use in dental applications that can benefit from 3D information, e.g., tooth angular misa-lignment detection and classification. Reconstructing 3D structures directly from 2D PX has recently been explored to address limitations with existing methods primarily reliant on Convolutional Neural Networks (CNNs) for direct 2D-to-3D mapping. These methods, however, are unable to correctly infer depth-axis spatial information. In addition, they are limited by the in-trinsic locality of convolution operations, as the convolution kernels only capture the information of immediate neighborhood pixels. In this study, we propose a progressive hybrid Multilayer Perceptron (MLP)-CNN pyra-mid network (3DPX) for 2D-to-3D oral PX reconstruction. We introduce a progressive reconstruction strategy, where 3D images are progressively re-constructed in the 3DPX with guidance imposed on the intermediate recon-struction result at each pyramid level. Further, motivated by the recent ad-vancement of MLPs that show promise in capturing fine-grained long-range dependency, our 3DPX integrates MLPs and CNNs to improve the semantic understanding during reconstruction. Extensive experiments on two large datasets involving 464 studies demonstrate that our 3DPX outperforms state-of-the-art 2D-to-3D oral reconstruction methods, including standalone MLP and transformers, in reconstruction quality, and also im-proves the performance of downstream angular misalignment classification tasks.
Correlation-aware Coarse-to-fine MLPs for Deformable Medical Image Registration
May 31, 2024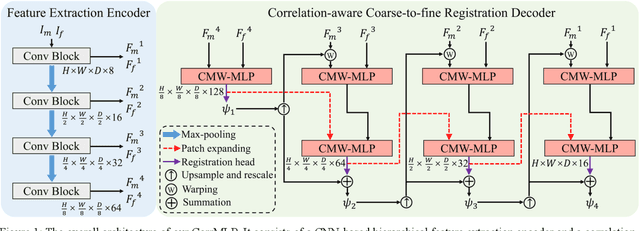
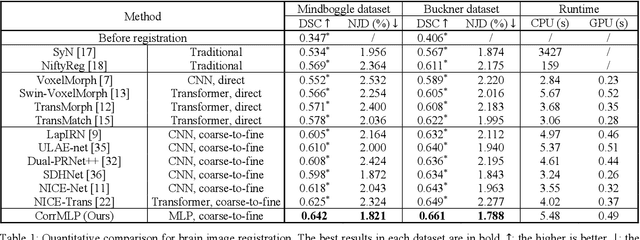
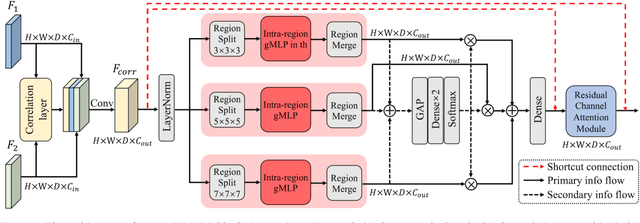
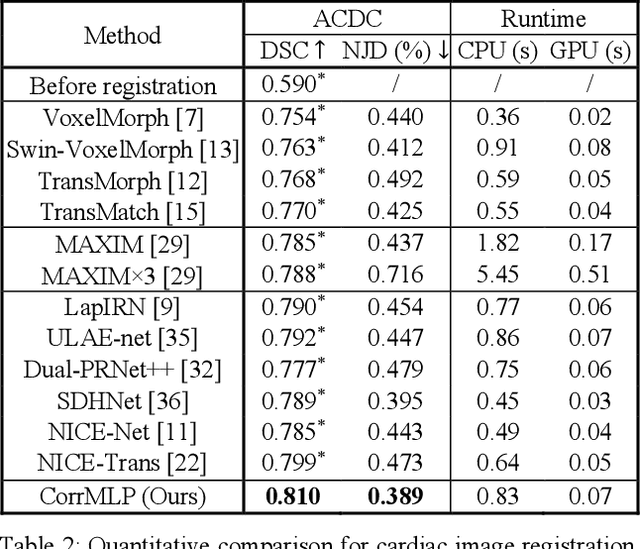
Abstract:Deformable image registration is a fundamental step for medical image analysis. Recently, transformers have been used for registration and outperformed Convolutional Neural Networks (CNNs). Transformers can capture long-range dependence among image features, which have been shown beneficial for registration. However, due to the high computation/memory loads of self-attention, transformers are typically used at downsampled feature resolutions and cannot capture fine-grained long-range dependence at the full image resolution. This limits deformable registration as it necessitates precise dense correspondence between each image pixel. Multi-layer Perceptrons (MLPs) without self-attention are efficient in computation/memory usage, enabling the feasibility of capturing fine-grained long-range dependence at full resolution. Nevertheless, MLPs have not been extensively explored for image registration and are lacking the consideration of inductive bias crucial for medical registration tasks. In this study, we propose the first correlation-aware MLP-based registration network (CorrMLP) for deformable medical image registration. Our CorrMLP introduces a correlation-aware multi-window MLP block in a novel coarse-to-fine registration architecture, which captures fine-grained multi-range dependence to perform correlation-aware coarse-to-fine registration. Extensive experiments with seven public medical datasets show that our CorrMLP outperforms state-of-the-art deformable registration methods.
Dual-modal Dynamic Traceback Learning for Medical Report Generation
Jan 24, 2024Abstract:With increasing reliance on medical imaging in clinical practices, automated report generation from medical images is in great demand. Existing report generation methods typically adopt an encoder-decoder deep learning framework to build a uni-directional image-to-report mapping. However, such a framework ignores the bi-directional mutual associations between images and reports, thus incurring difficulties in associating the intrinsic medical meanings between them. Recent generative representation learning methods have demonstrated the benefits of dual-modal learning from both image and text modalities. However, these methods exhibit two major drawbacks for medical report generation: 1) they tend to capture morphological information and have difficulties in capturing subtle pathological semantic information, and 2) they predict masked text rely on both unmasked images and text, inevitably degrading performance when inference is based solely on images. In this study, we propose a new report generation framework with dual-modal dynamic traceback learning (DTrace) to overcome the two identified drawbacks and enable dual-modal learning for medical report generation. To achieve this, our DTrace introduces a traceback mechanism to control the semantic validity of generated content via self-assessment. Further, our DTrace introduces a dynamic learning strategy to adapt to various proportions of image and text input, enabling report generation without reliance on textual input during inference. Extensive experiments on two well-benchmarked datasets (IU-Xray and MIMIC-CXR) show that our DTrace outperforms state-of-the-art medical report generation methods.
Full-resolution MLPs Empower Medical Dense Prediction
Nov 28, 2023



Abstract:Dense prediction is a fundamental requirement for many medical vision tasks such as medical image restoration, registration, and segmentation. The most popular vision model, Convolutional Neural Networks (CNNs), has reached bottlenecks due to the intrinsic locality of convolution operations. Recently, transformers have been widely adopted for dense prediction for their capability to capture long-range visual dependence. However, due to the high computational complexity and large memory consumption of self-attention operations, transformers are usually used at downsampled feature resolutions. Such usage cannot effectively leverage the tissue-level textural information available only at the full image resolution. This textural information is crucial for medical dense prediction as it can differentiate the subtle human anatomy in medical images. In this study, we hypothesize that Multi-layer Perceptrons (MLPs) are superior alternatives to transformers in medical dense prediction where tissue-level details dominate the performance, as MLPs enable long-range dependence at the full image resolution. To validate our hypothesis, we develop a full-resolution hierarchical MLP framework that uses MLPs beginning from the full image resolution. We evaluate this framework with various MLP blocks on a wide range of medical dense prediction tasks including restoration, registration, and segmentation. Extensive experiments on six public well-benchmarked datasets show that, by simply using MLPs at full resolution, our framework outperforms its CNN and transformer counterparts and achieves state-of-the-art performance on various medical dense prediction tasks.
AutoFuse: Automatic Fusion Networks for Deformable Medical Image Registration
Sep 11, 2023



Abstract:Deformable image registration aims to find a dense non-linear spatial correspondence between a pair of images, which is a crucial step for many medical tasks such as tumor growth monitoring and population analysis. Recently, Deep Neural Networks (DNNs) have been widely recognized for their ability to perform fast end-to-end registration. However, DNN-based registration needs to explore the spatial information of each image and fuse this information to characterize spatial correspondence. This raises an essential question: what is the optimal fusion strategy to characterize spatial correspondence? Existing fusion strategies (e.g., early fusion, late fusion) were empirically designed to fuse information by manually defined prior knowledge, which inevitably constrains the registration performance within the limits of empirical designs. In this study, we depart from existing empirically-designed fusion strategies and develop a data-driven fusion strategy for deformable image registration. To achieve this, we propose an Automatic Fusion network (AutoFuse) that provides flexibility to fuse information at many potential locations within the network. A Fusion Gate (FG) module is also proposed to control how to fuse information at each potential network location based on training data. Our AutoFuse can automatically optimize its fusion strategy during training and can be generalizable to both unsupervised registration (without any labels) and semi-supervised registration (with weak labels provided for partial training data). Extensive experiments on two well-benchmarked medical registration tasks (inter- and intra-patient registration) with eight public datasets show that our AutoFuse outperforms state-of-the-art unsupervised and semi-supervised registration methods.
Merging-Diverging Hybrid Transformer Networks for Survival Prediction in Head and Neck Cancer
Jul 07, 2023Abstract:Survival prediction is crucial for cancer patients as it provides early prognostic information for treatment planning. Recently, deep survival models based on deep learning and medical images have shown promising performance for survival prediction. However, existing deep survival models are not well developed in utilizing multi-modality images (e.g., PET-CT) and in extracting region-specific information (e.g., the prognostic information in Primary Tumor (PT) and Metastatic Lymph Node (MLN) regions). In view of this, we propose a merging-diverging learning framework for survival prediction from multi-modality images. This framework has a merging encoder to fuse multi-modality information and a diverging decoder to extract region-specific information. In the merging encoder, we propose a Hybrid Parallel Cross-Attention (HPCA) block to effectively fuse multi-modality features via parallel convolutional layers and cross-attention transformers. In the diverging decoder, we propose a Region-specific Attention Gate (RAG) block to screen out the features related to lesion regions. Our framework is demonstrated on survival prediction from PET-CT images in Head and Neck (H&N) cancer, by designing an X-shape merging-diverging hybrid transformer network (named XSurv). Our XSurv combines the complementary information in PET and CT images and extracts the region-specific prognostic information in PT and MLN regions. Extensive experiments on the public dataset of HEad and neCK TumOR segmentation and outcome prediction challenge (HECKTOR 2022) demonstrate that our XSurv outperforms state-of-the-art survival prediction methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge