Jinman Kim
HyperWalker: Dynamic Hypergraph-Based Deep Diagnosis for Multi-Hop Clinical Modeling across EHR and X-Ray in Medical VLMs
Jan 20, 2026Abstract:Automated clinical diagnosis remains a core challenge in medical AI, which usually requires models to integrate multi-modal data and reason across complex, case-specific contexts. Although recent methods have advanced medical report generation (MRG) and visual question answering (VQA) with medical vision-language models (VLMs), these methods, however, predominantly operate under a sample-isolated inference paradigm, as such processing cases independently without access to longitudinal electronic health records (EHRs) or structurally related patient examples. This paradigm limits reasoning to image-derived information alone, which ignores external complementary medical evidence for potentially more accurate diagnosis. To overcome this limitation, we propose \textbf{HyperWalker}, a \textit{Deep Diagnosis} framework that reformulates clinical reasoning via dynamic hypergraphs and test-time training. First, we construct a dynamic hypergraph, termed \textbf{iBrochure}, to model the structural heterogeneity of EHR data and implicit high-order associations among multimodal clinical information. Within this hypergraph, a reinforcement learning agent, \textbf{Walker}, navigates to and identifies optimal diagnostic paths. To ensure comprehensive coverage of diverse clinical characteristics in test samples, we incorporate a \textit{linger mechanism}, a multi-hop orthogonal retrieval strategy that iteratively selects clinically complementary neighborhood cases reflecting distinct clinical attributes. Experiments on MRG with MIMIC and medical VQA on EHRXQA demonstrate that HyperWalker achieves state-of-the-art performance. Code is available at: https://github.com/Bean-Young/HyperWalker
Intersectional Fairness in Vision-Language Models for Medical Image Disease Classification
Dec 24, 2025Abstract:Medical artificial intelligence (AI) systems, particularly multimodal vision-language models (VLM), often exhibit intersectional biases where models are systematically less confident in diagnosing marginalised patient subgroups. Such bias can lead to higher rates of inaccurate and missed diagnoses due to demographically skewed data and divergent distributions of diagnostic certainty. Current fairness interventions frequently fail to address these gaps or compromise overall diagnostic performance to achieve statistical parity among the subgroups. In this study, we developed Cross-Modal Alignment Consistency (CMAC-MMD), a training framework that standardises diagnostic certainty across intersectional patient subgroups. Unlike traditional debiasing methods, this approach equalises the model's decision confidence without requiring sensitive demographic data during clinical inference. We evaluated this approach using 10,015 skin lesion images (HAM10000) with external validation on 12,000 images (BCN20000), and 10,000 fundus images for glaucoma detection (Harvard-FairVLMed), stratifying performance by intersectional age, gender, and race attributes. In the dermatology cohort, the proposed method reduced the overall intersectional missed diagnosis gap (difference in True Positive Rate, $Δ$TPR) from 0.50 to 0.26 while improving the overall Area Under the Curve (AUC) from 0.94 to 0.97 compared to standard training. Similarly, for glaucoma screening, the method reduced $Δ$TPR from 0.41 to 0.31, achieving a better AUC of 0.72 (vs. 0.71 baseline). This establishes a scalable framework for developing high-stakes clinical decision support systems that are both accurate and can perform equitably across diverse patient subgroups, ensuring reliable performance without increasing privacy risks.
MRG-R1: Reinforcement Learning for Clinically Aligned Medical Report Generation
Dec 18, 2025Abstract:Medical report generation (MRG) aims to automatically derive radiology-style reports from medical images to aid in clinical decision-making. However, existing methods often generate text that mimics the linguistic style of radiologists but fails to guarantee clinical correctness, because they are trained on token-level objectives which focus on word-choice and sentence structure rather than actual medical accuracy. We propose a semantic-driven reinforcement learning (SRL) method for medical report generation, adopted on a large vision-language model (LVLM). SRL adopts Group Relative Policy Optimization (GRPO) to encourage clinical-correctness-guided learning beyond imitation of language style. Specifically, we optimise a report-level reward: a margin-based cosine similarity (MCCS) computed between key radiological findings extracted from generated and reference reports, thereby directly aligning clinical-label agreement and improving semantic correctness. A lightweight reasoning format constraint further guides the model to generate structured "thinking report" outputs. We evaluate Medical Report Generation with Sematic-driven Reinforment Learning (MRG-R1), on two datasets: IU X-Ray and MIMIC-CXR using clinical efficacy (CE) metrics. MRG-R1 achieves state-of-the-art performance with CE-F1 51.88 on IU X-Ray and 40.39 on MIMIC-CXR. We found that the label-semantic reinforcement is better than conventional token-level supervision. These results indicate that optimizing a clinically grounded, report-level reward rather than token overlap,meaningfully improves clinical correctness. This work is a prior to explore semantic-reinforcement in supervising medical correctness in medical Large vision-language model(Med-LVLM) training.
MRGAgents: A Multi-Agent Framework for Improved Medical Report Generation with Med-LVLMs
May 24, 2025Abstract:Medical Large Vision-Language Models (Med-LVLMs) have been widely adopted for medical report generation. Despite Med-LVLMs producing state-of-the-art performance, they exhibit a bias toward predicting all findings as normal, leading to reports that overlook critical abnormalities. Furthermore, these models often fail to provide comprehensive descriptions of radiologically relevant regions necessary for accurate diagnosis. To address these challenges, we proposeMedical Report Generation Agents (MRGAgents), a novel multi-agent framework that fine-tunes specialized agents for different disease categories. By curating subsets of the IU X-ray and MIMIC-CXR datasets to train disease-specific agents, MRGAgents generates reports that more effectively balance normal and abnormal findings while ensuring a comprehensive description of clinically relevant regions. Our experiments demonstrate that MRGAgents outperformed the state-of-the-art, improving both report comprehensiveness and diagnostic utility.
MedCFVQA: A Causal Approach to Mitigate Modality Preference Bias in Medical Visual Question Answering
May 23, 2025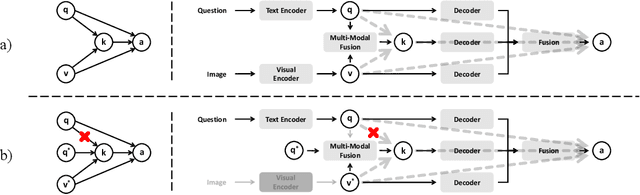
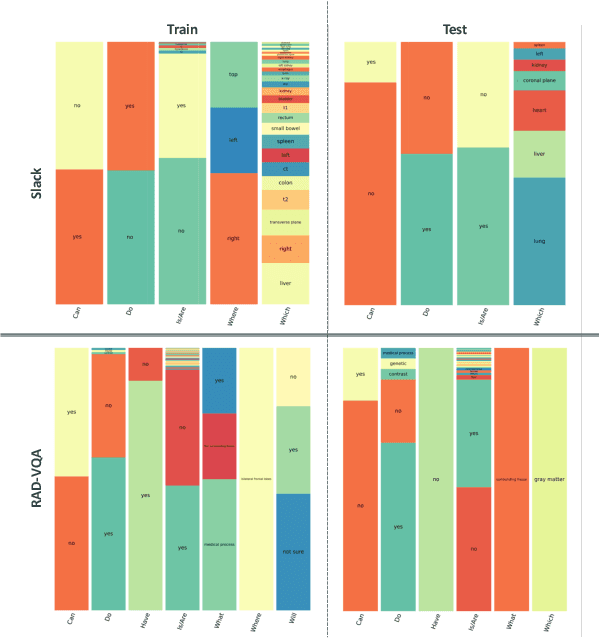
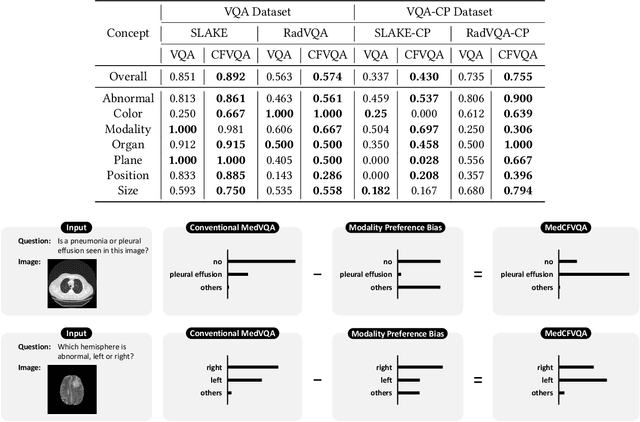
Abstract:Medical Visual Question Answering (MedVQA) is crucial for enhancing the efficiency of clinical diagnosis by providing accurate and timely responses to clinicians' inquiries regarding medical images. Existing MedVQA models suffered from modality preference bias, where predictions are heavily dominated by one modality while overlooking the other (in MedVQA, usually questions dominate the answer but images are overlooked), thereby failing to learn multimodal knowledge. To overcome the modality preference bias, we proposed a Medical CounterFactual VQA (MedCFVQA) model, which trains with bias and leverages causal graphs to eliminate the modality preference bias during inference. Existing MedVQA datasets exhibit substantial prior dependencies between questions and answers, which results in acceptable performance even if the model significantly suffers from the modality preference bias. To address this issue, we reconstructed new datasets by leveraging existing MedVQA datasets and Changed their P3rior dependencies (CP) between questions and their answers in the training and test set. Extensive experiments demonstrate that MedCFVQA significantly outperforms its non-causal counterpart on both SLAKE, RadVQA and SLAKE-CP, RadVQA-CP datasets.
A Causal Approach to Mitigate Modality Preference Bias in Medical Visual Question Answering
May 22, 2025


Abstract:Medical Visual Question Answering (MedVQA) is crucial for enhancing the efficiency of clinical diagnosis by providing accurate and timely responses to clinicians' inquiries regarding medical images. Existing MedVQA models suffered from modality preference bias, where predictions are heavily dominated by one modality while overlooking the other (in MedVQA, usually questions dominate the answer but images are overlooked), thereby failing to learn multimodal knowledge. To overcome the modality preference bias, we proposed a Medical CounterFactual VQA (MedCFVQA) model, which trains with bias and leverages causal graphs to eliminate the modality preference bias during inference. Existing MedVQA datasets exhibit substantial prior dependencies between questions and answers, which results in acceptable performance even if the model significantly suffers from the modality preference bias. To address this issue, we reconstructed new datasets by leveraging existing MedVQA datasets and Changed their P3rior dependencies (CP) between questions and their answers in the training and test set. Extensive experiments demonstrate that MedCFVQA significantly outperforms its non-causal counterpart on both SLAKE, RadVQA and SLAKE-CP, RadVQA-CP datasets.
MAISY: Motion-Aware Image SYnthesis for Medical Image Motion Correction
May 08, 2025Abstract:Patient motion during medical image acquisition causes blurring, ghosting, and distorts organs, which makes image interpretation challenging. Current state-of-the-art algorithms using Generative Adversarial Network (GAN)-based methods with their ability to learn the mappings between corrupted images and their ground truth via Structural Similarity Index Measure (SSIM) loss effectively generate motion-free images. However, we identified the following limitations: (i) they mainly focus on global structural characteristics and therefore overlook localized features that often carry critical pathological information, and (ii) the SSIM loss function struggles to handle images with varying pixel intensities, luminance factors, and variance. In this study, we propose Motion-Aware Image SYnthesis (MAISY) which initially characterize motion and then uses it for correction by: (a) leveraging the foundation model Segment Anything Model (SAM), to dynamically learn spatial patterns along anatomical boundaries where motion artifacts are most pronounced and, (b) introducing the Variance-Selective SSIM (VS-SSIM) loss which adaptively emphasizes spatial regions with high pixel variance to preserve essential anatomical details during artifact correction. Experiments on chest and head CT datasets demonstrate that our model outperformed the state-of-the-art counterparts, with Peak Signal-to-Noise Ratio (PSNR) increasing by 40%, SSIM by 10%, and Dice by 16%.
SMPL-GPTexture: Dual-View 3D Human Texture Estimation using Text-to-Image Generation Models
Apr 17, 2025Abstract:Generating high-quality, photorealistic textures for 3D human avatars remains a fundamental yet challenging task in computer vision and multimedia field. However, real paired front and back images of human subjects are rarely available with privacy, ethical and cost of acquisition, which restricts scalability of the data. Additionally, learning priors from image inputs using deep generative models, such as GANs or diffusion models, to infer unseen regions such as the human back often leads to artifacts, structural inconsistencies, or loss of fine-grained detail. To address these issues, we present SMPL-GPTexture (skinned multi-person linear model - general purpose Texture), a novel pipeline that takes natural language prompts as input and leverages a state-of-the-art text-to-image generation model to produce paired high-resolution front and back images of a human subject as the starting point for texture estimation. Using the generated paired dual-view images, we first employ a human mesh recovery model to obtain a robust 2D-to-3D SMPL alignment between image pixels and the 3D model's UV coordinates for each views. Second, we use an inverted rasterization technique that explicitly projects the observed colour from the input images into the UV space, thereby producing accurate, complete texture maps. Finally, we apply a diffusion-based inpainting module to fill in the missing regions, and the fusion mechanism then combines these results into a unified full texture map. Extensive experiments shows that our SMPL-GPTexture can generate high resolution texture aligned with user's prompts.
Bridging the Semantic Gaps: Improving Medical VQA Consistency with LLM-Augmented Question Sets
Apr 16, 2025Abstract:Medical Visual Question Answering (MVQA) systems can interpret medical images in response to natural language queries. However, linguistic variability in question phrasing often undermines the consistency of these systems. To address this challenge, we propose a Semantically Equivalent Question Augmentation (SEQA) framework, which leverages large language models (LLMs) to generate diverse yet semantically equivalent rephrasings of questions. Specifically, this approach enriches linguistic diversity while preserving semantic meaning. We further introduce an evaluation metric, Total Agreement Rate with Semantically Equivalent Input and Correct Answer (TAR-SC), which assesses a model's capability to generate consistent and correct responses to semantically equivalent linguistic variations. In addition, we also propose three other diversity metrics - average number of QA items per image (ANQI), average number of questions per image with the same answer (ANQA), and average number of open-ended questions per image with the same semantics (ANQS). Using the SEQA framework, we augmented the benchmarked MVQA public datasets of SLAKE, VQA-RAD, and PathVQA. As a result, all three datasets achieved significant improvements by incorporating more semantically equivalent questions: ANQI increased by an average of 86.1, ANQA by 85.1, and ANQS by 46. Subsequent experiments evaluate three MVQA models (M2I2, MUMC, and BiomedGPT) under both zero-shot and fine-tuning settings on the enhanced datasets. Experimental results in MVQA datasets show that fine-tuned models achieve an average accuracy improvement of 19.35%, while our proposed TAR-SC metric shows an average improvement of 11. 61%, indicating a substantial enhancement in model consistency.
Multi-modal 3D Pose and Shape Estimation with Computed Tomography
Mar 25, 2025



Abstract:In perioperative care, precise in-bed 3D patient pose and shape estimation (PSE) can be vital in optimizing patient positioning in preoperative planning, enabling accurate overlay of medical images for augmented reality-based surgical navigation, and mitigating risks of prolonged immobility during recovery. Conventional PSE methods relying on modalities such as RGB-D, infrared, or pressure maps often struggle with occlusions caused by bedding and complex patient positioning, leading to inaccurate estimation that can affect clinical outcomes. To address these challenges, we present the first multi-modal in-bed patient 3D PSE network that fuses detailed geometric features extracted from routinely acquired computed tomography (CT) scans with depth maps (mPSE-CT). mPSE-CT incorporates a shape estimation module that utilizes probabilistic correspondence alignment, a pose estimation module with a refined neural network, and a final parameters mixing module. This multi-modal network robustly reconstructs occluded body regions and enhances the accuracy of the estimated 3D human mesh model. We validated mPSE-CT using proprietary whole-body rigid phantom and volunteer datasets in clinical scenarios. mPSE-CT outperformed the best-performing prior method by 23% and 49.16% in pose and shape estimation respectively, demonstrating its potential for improving clinical outcomes in challenging perioperative environments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge