Lei Bi
HyperWalker: Dynamic Hypergraph-Based Deep Diagnosis for Multi-Hop Clinical Modeling across EHR and X-Ray in Medical VLMs
Jan 20, 2026Abstract:Automated clinical diagnosis remains a core challenge in medical AI, which usually requires models to integrate multi-modal data and reason across complex, case-specific contexts. Although recent methods have advanced medical report generation (MRG) and visual question answering (VQA) with medical vision-language models (VLMs), these methods, however, predominantly operate under a sample-isolated inference paradigm, as such processing cases independently without access to longitudinal electronic health records (EHRs) or structurally related patient examples. This paradigm limits reasoning to image-derived information alone, which ignores external complementary medical evidence for potentially more accurate diagnosis. To overcome this limitation, we propose \textbf{HyperWalker}, a \textit{Deep Diagnosis} framework that reformulates clinical reasoning via dynamic hypergraphs and test-time training. First, we construct a dynamic hypergraph, termed \textbf{iBrochure}, to model the structural heterogeneity of EHR data and implicit high-order associations among multimodal clinical information. Within this hypergraph, a reinforcement learning agent, \textbf{Walker}, navigates to and identifies optimal diagnostic paths. To ensure comprehensive coverage of diverse clinical characteristics in test samples, we incorporate a \textit{linger mechanism}, a multi-hop orthogonal retrieval strategy that iteratively selects clinically complementary neighborhood cases reflecting distinct clinical attributes. Experiments on MRG with MIMIC and medical VQA on EHRXQA demonstrate that HyperWalker achieves state-of-the-art performance. Code is available at: https://github.com/Bean-Young/HyperWalker
Advancing Deformable Medical Image Registration with Multi-axis Cross-covariance Attention
Dec 24, 2024Abstract:Deformable image registration is a fundamental requirement for medical image analysis. Recently, transformers have been widely used in deep learning-based registration methods for their ability to capture long-range dependency via self-attention (SA). However, the high computation and memory loads of SA (growing quadratically with the spatial resolution) hinder transformers from processing subtle textural information in high-resolution image features, e.g., at the full and half image resolutions. This limits deformable registration as the high-resolution textural information is crucial for finding precise pixel-wise correspondence between subtle anatomical structures. Cross-covariance Attention (XCA), as a "transposed" version of SA that operates across feature channels, has complexity growing linearly with the spatial resolution, providing the feasibility of capturing long-range dependency among high-resolution image features. However, existing XCA-based transformers merely capture coarse global long-range dependency, which are unsuitable for deformable image registration relying primarily on fine-grained local correspondence. In this study, we propose to improve existing deep learning-based registration methods by embedding a new XCA mechanism. To this end, we design an XCA-based transformer block optimized for deformable medical image registration, named Multi-Axis XCA (MAXCA). Our MAXCA serves as a general network block that can be embedded into various registration network architectures. It can capture both global and local long-range dependency among high-resolution image features by applying regional and dilated XCA in parallel via a multi-axis design. Extensive experiments on two well-benchmarked inter-/intra-patient registration tasks with seven public medical datasets demonstrate that our MAXCA block enables state-of-the-art registration performance.
Language-guided Medical Image Segmentation with Target-informed Multi-level Contrastive Alignments
Dec 18, 2024



Abstract:Medical image segmentation is crucial in modern medical image analysis, which can aid into diagnosis of various disease conditions. Recently, language-guided segmentation methods have shown promising results in automating image segmentation where text reports are incorporated as guidance. These text reports, containing image impressions and insights given by clinicians, provides auxiliary guidance. However, these methods neglect the inherent pattern gaps between the two distinct modalities, which leads to sub-optimal image-text feature fusion without proper cross-modality feature alignments. Contrastive alignments are widely used to associate image-text semantics in representation learning; however, it has not been exploited to bridge the pattern gaps in language-guided segmentation that relies on subtle low level image details to represent diseases. Existing contrastive alignment methods typically algin high-level global image semantics without involving low-level, localized target information, and therefore fails to explore fine-grained text guidance for language-guided segmentation. In this study, we propose a language-guided segmentation network with Target-informed Multi-level Contrastive Alignments (TMCA). TMCA enables target-informed cross-modality alignments and fine-grained text guidance to bridge the pattern gaps in language-guided segmentation. Specifically, we introduce: 1) a target-sensitive semantic distance module that enables granular image-text alignment modelling, and 2) a multi-level alignment strategy that directs text guidance on low-level image features. In addition, a language-guided target enhancement module is proposed to leverage the aligned text to redirect attention to focus on critical localized image features. Extensive experiments on 4 image-text datasets, involving 3 medical imaging modalities, demonstrated that our TMCA achieved superior performances.
Automatic Left Ventricular Cavity Segmentation via Deep Spatial Sequential Network in 4D Computed Tomography Studies
Dec 17, 2024



Abstract:Automated segmentation of left ventricular cavity (LVC) in temporal cardiac image sequences (multiple time points) is a fundamental requirement for quantitative analysis of its structural and functional changes. Deep learning based methods for the segmentation of LVC are the state of the art; however, these methods are generally formulated to work on single time points, and fails to exploit the complementary information from the temporal image sequences that can aid in segmentation accuracy and consistency among the images across the time points. Furthermore, these segmentation methods perform poorly in segmenting the end-systole (ES) phase images, where the left ventricle deforms to the smallest irregular shape, and the boundary between the blood chamber and myocardium becomes inconspicuous. To overcome these limitations, we propose a new method to automatically segment temporal cardiac images where we introduce a spatial sequential (SS) network to learn the deformation and motion characteristics of the LVC in an unsupervised manner; these characteristics were then integrated with sequential context information derived from bi-directional learning (BL) where both chronological and reverse-chronological directions of the image sequence were used. Our experimental results on a cardiac computed tomography (CT) dataset demonstrated that our spatial-sequential network with bi-directional learning (SS-BL) method outperformed existing methods for LVC segmentation. Our method was also applied to MRI cardiac dataset and the results demonstrated the generalizability of our method.
Hyper-Fusion Network for Semi-Automatic Segmentation of Skin Lesions
Dec 14, 2024Abstract:Automatic skin lesion segmentation methods based on fully convolutional networks (FCNs) are regarded as the state-of-the-art for accuracy. When there are, however, insufficient training data to cover all the variations in skin lesions, where lesions from different patients may have major differences in size/shape/texture, these methods failed to segment the lesions that have image characteristics, which are less common in the training datasets. FCN-based semi-automatic segmentation methods, which fuse user-inputs with high-level semantic image features derived from FCNs offer an ideal complement to overcome limitations of automatic segmentation methods. These semi-automatic methods rely on the automated state-of-the-art FCNs coupled with user-inputs for refinements, and therefore being able to tackle challenging skin lesions. However, there are a limited number of FCN-based semi-automatic segmentation methods and all these methods focused on early-fusion, where the first few convolutional layers are used to fuse image features and user-inputs and then derive fused image features for segmentation. For early-fusion based methods, because the user-input information can be lost after the first few convolutional layers, consequently, the user-input information will have limited guidance and constraint in segmenting the challenging skin lesions with inhomogeneous textures and fuzzy boundaries. Hence, in this work, we introduce a hyper-fusion network (HFN) to fuse the extracted user-inputs and image features over multiple stages. We separately extract complementary features which then allows for an iterative use of user-inputs along all the fusion stages to refine the segmentation. We evaluated our HFN on ISIC 2017, ISIC 2016 and PH2 datasets, and our results show that the HFN is more accurate and generalizable than the state-of-the-art methods.
3DPX: Single Panoramic X-ray Analysis Guided by 3D Oral Structure Reconstruction
Sep 27, 2024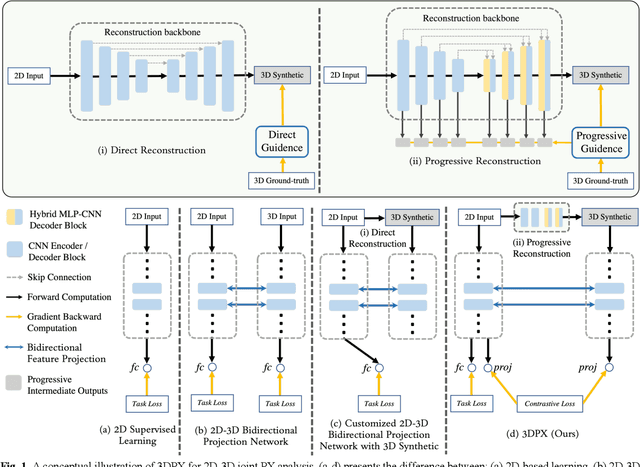
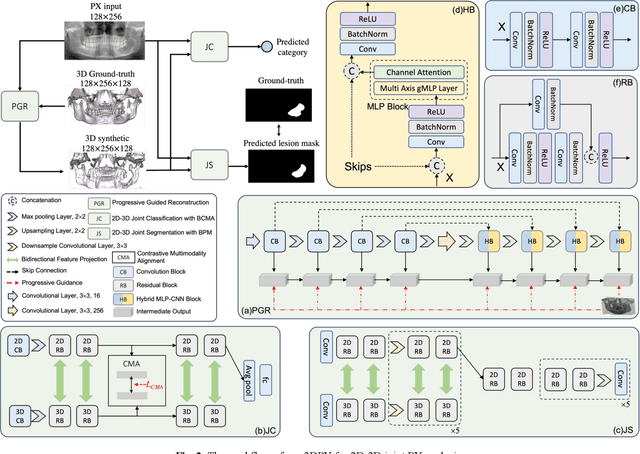
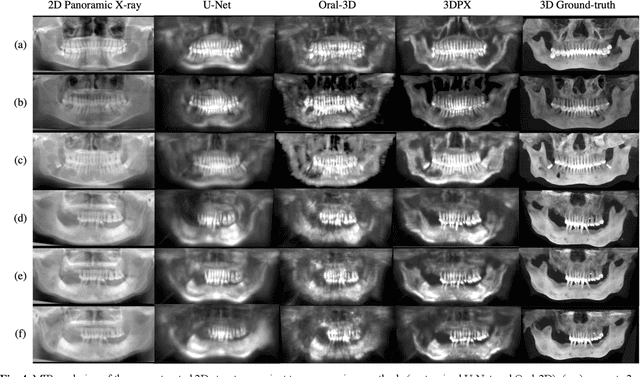
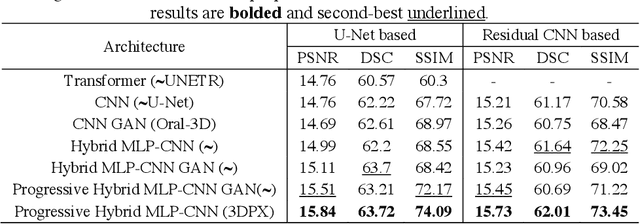
Abstract:Panoramic X-ray (PX) is a prevalent modality in dentistry practice owing to its wide availability and low cost. However, as a 2D projection of a 3D structure, PX suffers from anatomical information loss and PX diagnosis is limited compared to that with 3D imaging modalities. 2D-to-3D reconstruction methods have been explored for the ability to synthesize the absent 3D anatomical information from 2D PX for use in PX image analysis. However, there are challenges in leveraging such 3D synthesized reconstructions. First, inferring 3D depth from 2D images remains a challenging task with limited accuracy. The second challenge is the joint analysis of 2D PX with its 3D synthesized counterpart, with the aim to maximize the 2D-3D synergy while minimizing the errors arising from the synthesized image. In this study, we propose a new method termed 3DPX - PX image analysis guided by 2D-to-3D reconstruction, to overcome these challenges. 3DPX consists of (i) a novel progressive reconstruction network to improve 2D-to-3D reconstruction and, (ii) a contrastive-guided bidirectional multimodality alignment module for 3D-guided 2D PX classification and segmentation tasks. The reconstruction network progressively reconstructs 3D images with knowledge imposed on the intermediate reconstructions at multiple pyramid levels and incorporates Multilayer Perceptrons to improve semantic understanding. The downstream networks leverage the reconstructed images as 3D anatomical guidance to the PX analysis through feature alignment, which increases the 2D-3D synergy with bidirectional feature projection and decease the impact of potential errors with contrastive guidance. Extensive experiments on two oral datasets involving 464 studies demonstrate that 3DPX outperforms the state-of-the-art methods in various tasks including 2D-to-3D reconstruction, PX classification and lesion segmentation.
3DPX: Progressive 2D-to-3D Oral Image Reconstruction with Hybrid MLP-CNN Networks
Aug 02, 2024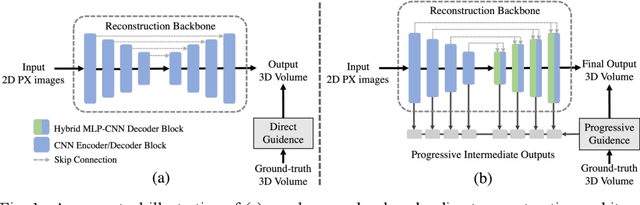
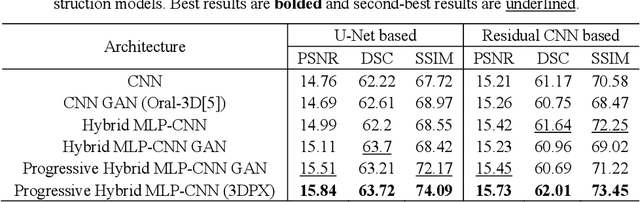
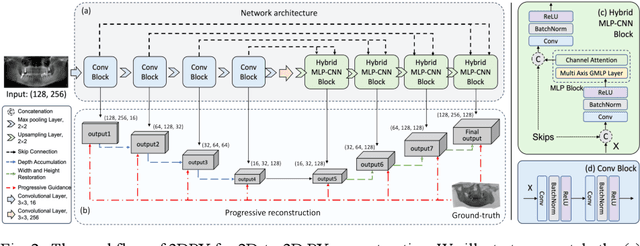
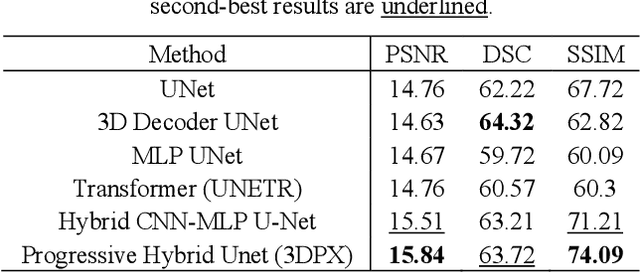
Abstract:Panoramic X-ray (PX) is a prevalent modality in dental practice for its wide availability and low cost. However, as a 2D projection image, PX does not contain 3D anatomical information, and therefore has limited use in dental applications that can benefit from 3D information, e.g., tooth angular misa-lignment detection and classification. Reconstructing 3D structures directly from 2D PX has recently been explored to address limitations with existing methods primarily reliant on Convolutional Neural Networks (CNNs) for direct 2D-to-3D mapping. These methods, however, are unable to correctly infer depth-axis spatial information. In addition, they are limited by the in-trinsic locality of convolution operations, as the convolution kernels only capture the information of immediate neighborhood pixels. In this study, we propose a progressive hybrid Multilayer Perceptron (MLP)-CNN pyra-mid network (3DPX) for 2D-to-3D oral PX reconstruction. We introduce a progressive reconstruction strategy, where 3D images are progressively re-constructed in the 3DPX with guidance imposed on the intermediate recon-struction result at each pyramid level. Further, motivated by the recent ad-vancement of MLPs that show promise in capturing fine-grained long-range dependency, our 3DPX integrates MLPs and CNNs to improve the semantic understanding during reconstruction. Extensive experiments on two large datasets involving 464 studies demonstrate that our 3DPX outperforms state-of-the-art 2D-to-3D oral reconstruction methods, including standalone MLP and transformers, in reconstruction quality, and also im-proves the performance of downstream angular misalignment classification tasks.
Correlation-aware Coarse-to-fine MLPs for Deformable Medical Image Registration
May 31, 2024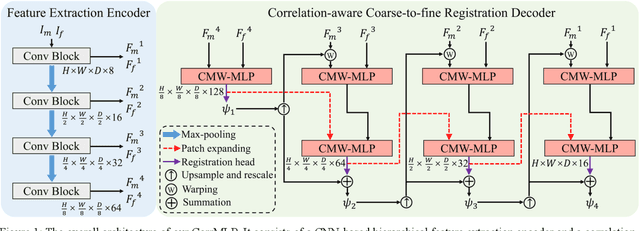
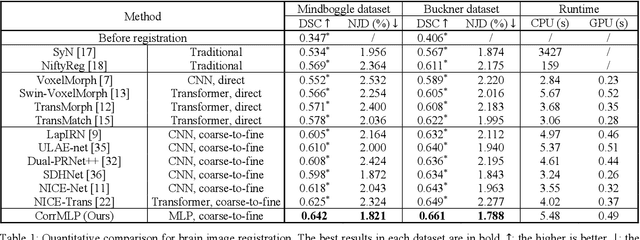
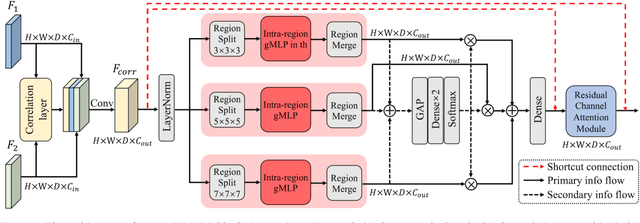
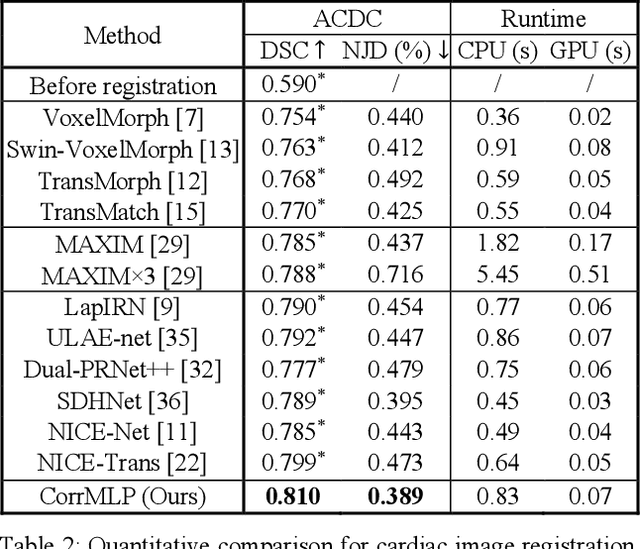
Abstract:Deformable image registration is a fundamental step for medical image analysis. Recently, transformers have been used for registration and outperformed Convolutional Neural Networks (CNNs). Transformers can capture long-range dependence among image features, which have been shown beneficial for registration. However, due to the high computation/memory loads of self-attention, transformers are typically used at downsampled feature resolutions and cannot capture fine-grained long-range dependence at the full image resolution. This limits deformable registration as it necessitates precise dense correspondence between each image pixel. Multi-layer Perceptrons (MLPs) without self-attention are efficient in computation/memory usage, enabling the feasibility of capturing fine-grained long-range dependence at full resolution. Nevertheless, MLPs have not been extensively explored for image registration and are lacking the consideration of inductive bias crucial for medical registration tasks. In this study, we propose the first correlation-aware MLP-based registration network (CorrMLP) for deformable medical image registration. Our CorrMLP introduces a correlation-aware multi-window MLP block in a novel coarse-to-fine registration architecture, which captures fine-grained multi-range dependence to perform correlation-aware coarse-to-fine registration. Extensive experiments with seven public medical datasets show that our CorrMLP outperforms state-of-the-art deformable registration methods.
Enhancing medical vision-language contrastive learning via inter-matching relation modelling
Jan 19, 2024Abstract:Medical image representations can be learned through medical vision-language contrastive learning (mVLCL) where medical imaging reports are used as weak supervision through image-text alignment. These learned image representations can be transferred to and benefit various downstream medical vision tasks such as disease classification and segmentation. Recent mVLCL methods attempt to align image sub-regions and the report keywords as local-matchings. However, these methods aggregate all local-matchings via simple pooling operations while ignoring the inherent relations between them. These methods therefore fail to reason between local-matchings that are semantically related, e.g., local-matchings that correspond to the disease word and the location word (semantic-relations), and also fail to differentiate such clinically important local-matchings from others that correspond to less meaningful words, e.g., conjunction words (importance-relations). Hence, we propose a mVLCL method that models the inter-matching relations between local-matchings via a relation-enhanced contrastive learning framework (RECLF). In RECLF, we introduce a semantic-relation reasoning module (SRM) and an importance-relation reasoning module (IRM) to enable more fine-grained report supervision for image representation learning. We evaluated our method using four public benchmark datasets on four downstream tasks, including segmentation, zero-shot classification, supervised classification, and cross-modal retrieval. Our results demonstrated the superiority of our RECLF over the state-of-the-art mVLCL methods with consistent improvements across single-modal and cross-modal tasks. These results suggest that our RECLF, by modelling the inter-matching relations, can learn improved medical image representations with better generalization capabilities.
Full-resolution MLPs Empower Medical Dense Prediction
Nov 28, 2023



Abstract:Dense prediction is a fundamental requirement for many medical vision tasks such as medical image restoration, registration, and segmentation. The most popular vision model, Convolutional Neural Networks (CNNs), has reached bottlenecks due to the intrinsic locality of convolution operations. Recently, transformers have been widely adopted for dense prediction for their capability to capture long-range visual dependence. However, due to the high computational complexity and large memory consumption of self-attention operations, transformers are usually used at downsampled feature resolutions. Such usage cannot effectively leverage the tissue-level textural information available only at the full image resolution. This textural information is crucial for medical dense prediction as it can differentiate the subtle human anatomy in medical images. In this study, we hypothesize that Multi-layer Perceptrons (MLPs) are superior alternatives to transformers in medical dense prediction where tissue-level details dominate the performance, as MLPs enable long-range dependence at the full image resolution. To validate our hypothesis, we develop a full-resolution hierarchical MLP framework that uses MLPs beginning from the full image resolution. We evaluate this framework with various MLP blocks on a wide range of medical dense prediction tasks including restoration, registration, and segmentation. Extensive experiments on six public well-benchmarked datasets show that, by simply using MLPs at full resolution, our framework outperforms its CNN and transformer counterparts and achieves state-of-the-art performance on various medical dense prediction tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge