Euijoon Ahn
Unsupervised Radar Point Cloud Enhancement via Arbitrary LiDAR Guided Diffusion Prior
May 15, 2025Abstract:In industrial automation, radar is a critical sensor in machine perception. However, the angular resolution of radar is inherently limited by the Rayleigh criterion, which depends on both the radar's operating wavelength and the effective aperture of its antenna array.To overcome these hardware-imposed limitations, recent neural network-based methods have leveraged high-resolution LiDAR data, paired with radar measurements, during training to enhance radar point cloud resolution. While effective, these approaches require extensive paired datasets, which are costly to acquire and prone to calibration error. These challenges motivate the need for methods that can improve radar resolution without relying on paired high-resolution ground-truth data. Here, we introduce an unsupervised radar points enhancement algorithm that employs an arbitrary LiDAR-guided diffusion model as a prior without the need for paired training data. Specifically, our approach formulates radar angle estimation recovery as an inverse problem and incorporates prior knowledge through a diffusion model with arbitrary LiDAR domain knowledge. Experimental results demonstrate that our method attains high fidelity and low noise performance compared to traditional regularization techniques. Additionally, compared to paired training methods, it not only achieves comparable performance but also offers improved generalization capability. To our knowledge, this is the first approach that enhances radar points output by integrating prior knowledge via a diffusion model rather than relying on paired training data. Our code is available at https://github.com/yyxr75/RadarINV.
Computationally Efficient Diffusion Models in Medical Imaging: A Comprehensive Review
May 09, 2025Abstract:The diffusion model has recently emerged as a potent approach in computer vision, demonstrating remarkable performances in the field of generative artificial intelligence. Capable of producing high-quality synthetic images, diffusion models have been successfully applied across a range of applications. However, a significant challenge remains with the high computational cost associated with training and generating these models. This study focuses on the efficiency and inference time of diffusion-based generative models, highlighting their applications in both natural and medical imaging. We present the most recent advances in diffusion models by categorizing them into three key models: the Denoising Diffusion Probabilistic Model (DDPM), the Latent Diffusion Model (LDM), and the Wavelet Diffusion Model (WDM). These models play a crucial role in medical imaging, where producing fast, reliable, and high-quality medical images is essential for accurate analysis of abnormalities and disease diagnosis. We first investigate the general framework of DDPM, LDM, and WDM and discuss the computational complexity gap filled by these models in natural and medical imaging. We then discuss the current limitations of these models as well as the opportunities and future research directions in medical imaging.
Improving Automatic Fetal Biometry Measurement with Swoosh Activation Function
Dec 16, 2024Abstract:The measurement of fetal thalamus diameter (FTD) and fetal head circumference (FHC) are crucial in identifying abnormal fetal thalamus development as it may lead to certain neuropsychiatric disorders in later life. However, manual measurements from 2D-US images are laborious, prone to high inter-observer variability, and complicated by the high signal-to-noise ratio nature of the images. Deep learning-based landmark detection approaches have shown promise in measuring biometrics from US images, but the current state-of-the-art (SOTA) algorithm, BiometryNet, is inadequate for FTD and FHC measurement due to its inability to account for the fuzzy edges of these structures and the complex shape of the FTD structure. To address these inadequacies, we propose a novel Swoosh Activation Function (SAF) designed to enhance the regularization of heatmaps produced by landmark detection algorithms. Our SAF serves as a regularization term to enforce an optimum mean squared error (MSE) level between predicted heatmaps, reducing the dispersiveness of hotspots in predicted heatmaps. Our experimental results demonstrate that SAF significantly improves the measurement performances of FTD and FHC with higher intraclass correlation coefficient scores in FTD and lower mean difference scores in FHC measurement than those of the current SOTA algorithm BiometryNet. Moreover, our proposed SAF is highly generalizable and architecture-agnostic. The SAF's coefficients can be configured for different tasks, making it highly customizable. Our study demonstrates that the SAF activation function is a novel method that can improve measurement accuracy in fetal biometry landmark detection. This improvement has the potential to contribute to better fetal monitoring and improved neonatal outcomes.
GVT2RPM: An Empirical Study for General Video Transformer Adaptation to Remote Physiological Measurement
Jun 19, 2024

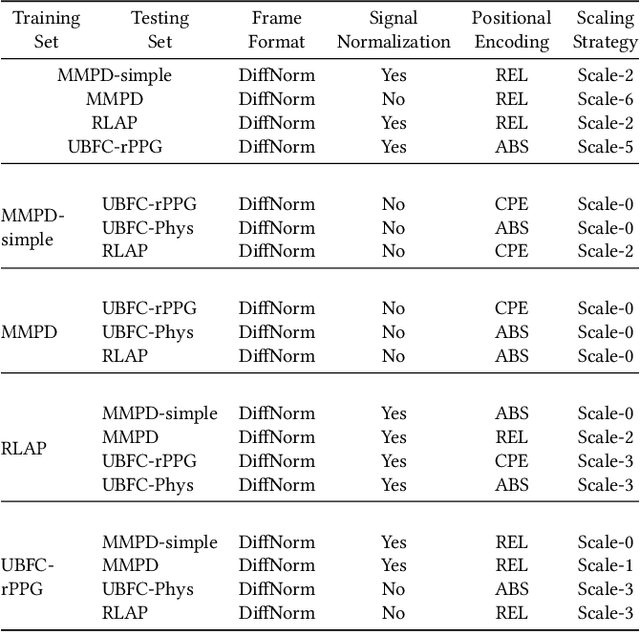

Abstract:Remote physiological measurement (RPM) is an essential tool for healthcare monitoring as it enables the measurement of physiological signs, e.g., heart rate, in a remote setting via physical wearables. Recently, with facial videos, we have seen rapid advancements in video-based RPMs. However, adopting facial videos for RPM in the clinical setting largely depends on the accuracy and robustness (work across patient populations). Fortunately, the capability of the state-of-the-art transformer architecture in general (natural) video understanding has resulted in marked improvements and has been translated to facial understanding, including RPM. However, existing RPM methods usually need RPM-specific modules, e.g., temporal difference convolution and handcrafted feature maps. Although these customized modules can increase accuracy, they are not demonstrated for their robustness across datasets. Further, due to their customization of the transformer architecture, they cannot use the advancements made in general video transformers (GVT). In this study, we interrogate the GVT architecture and empirically analyze how the training designs, i.e., data pre-processing and network configurations, affect the model performance applied to RPM. Based on the structure of video transformers, we propose to configure its spatiotemporal hierarchy to align with the dense temporal information needed in RPM for signal feature extraction. We define several practical guidelines and gradually adapt GVTs for RPM without introducing RPM-specific modules. Our experiments demonstrate favorable results to existing RPM-specific module counterparts. We conducted extensive experiments with five datasets using intra-dataset and cross-dataset settings. We highlight that the proposed guidelines GVT2RPM can be generalized to any video transformers and is robust to various datasets.
Self-Supervised Multi-Modality Learning for Multi-Label Skin Lesion Classification
Oct 28, 2023Abstract:The clinical diagnosis of skin lesion involves the analysis of dermoscopic and clinical modalities. Dermoscopic images provide a detailed view of the surface structures whereas clinical images offer a complementary macroscopic information. The visual diagnosis of melanoma is also based on seven-point checklist which involves identifying different visual attributes. Recently, supervised learning approaches such as convolutional neural networks (CNNs) have shown great performances using both dermoscopic and clinical modalities (Multi-modality). The seven different visual attributes in the checklist are also used to further improve the the diagnosis. The performances of these approaches, however, are still reliant on the availability of large-scaled labeled data. The acquisition of annotated dataset is an expensive and time-consuming task, more so with annotating multi-attributes. To overcome this limitation, we propose a self-supervised learning (SSL) algorithm for multi-modality skin lesion classification. Our algorithm enables the multi-modality learning by maximizing the similarities between paired dermoscopic and clinical images from different views. In addition, we generate surrogate pseudo-multi-labels that represent seven attributes via clustering analysis. We also propose a label-relation-aware module to refine each pseudo-label embedding and capture the interrelationships between pseudo-multi-labels. We validated the effectiveness of our algorithm using well-benchmarked seven-point skin lesion dataset. Our results show that our algorithm achieved better performances than other state-of-the-art SSL counterparts.
A Dual-branch Self-supervised Representation Learning Framework for Tumour Segmentation in Whole Slide Images
Mar 20, 2023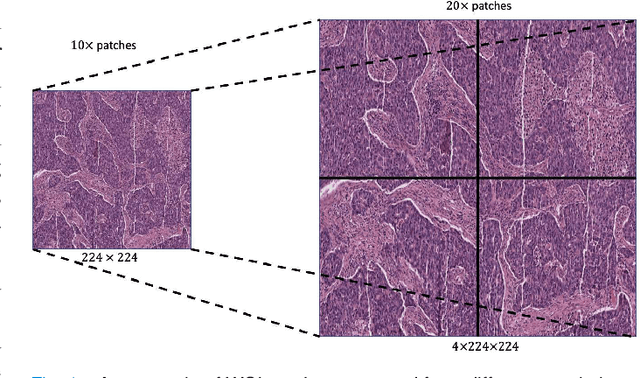
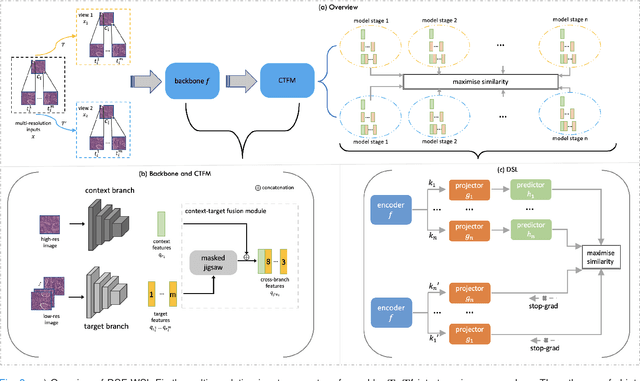
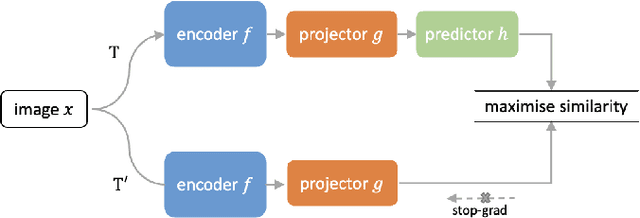
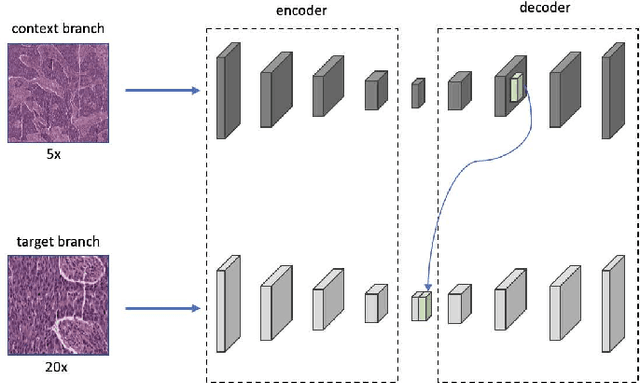
Abstract:Supervised deep learning methods have achieved considerable success in medical image analysis, owing to the availability of large-scale and well-annotated datasets. However, creating such datasets for whole slide images (WSIs) in histopathology is a challenging task due to their gigapixel size. In recent years, self-supervised learning (SSL) has emerged as an alternative solution to reduce the annotation overheads in WSIs, as it does not require labels for training. These SSL approaches, however, are not designed for handling multi-resolution WSIs, which limits their performance in learning discriminative image features. In this paper, we propose a Dual-branch SSL Framework for WSI tumour segmentation (DSF-WSI) that can effectively learn image features from multi-resolution WSIs. Our DSF-WSI connected two branches and jointly learnt low and high resolution WSIs in a self-supervised manner. Moreover, we introduced a novel Context-Target Fusion Module (CTFM) and a masked jigsaw pretext task to align the learnt multi-resolution features. Furthermore, we designed a Dense SimSiam Learning (DSL) strategy to maximise the similarity of different views of WSIs, enabling the learnt representations to be more efficient and discriminative. We evaluated our method using two public datasets on breast and liver cancer segmentation tasks. The experiment results demonstrated that our DSF-WSI can effectively extract robust and efficient representations, which we validated through subsequent fine-tuning and semi-supervised settings. Our proposed method achieved better accuracy than other state-of-the-art approaches. Code is available at https://github.com/Dylan-H-Wang/dsf-wsi.
A Review of Predictive and Contrastive Self-supervised Learning for Medical Images
Feb 10, 2023Abstract:Over the last decade, supervised deep learning on manually annotated big data has been progressing significantly on computer vision tasks. But the application of deep learning in medical image analysis was limited by the scarcity of high-quality annotated medical imaging data. An emerging solution is self-supervised learning (SSL), among which contrastive SSL is the most successful approach to rivalling or outperforming supervised learning. This review investigates several state-of-the-art contrastive SSL algorithms originally on natural images as well as their adaptations for medical images, and concludes by discussing recent advances, current limitations, and future directions in applying contrastive SSL in the medical domain.
Z-SSMNet: A Zonal-aware Self-Supervised Mesh Network for Prostate Cancer Detection and Diagnosis in bpMRI
Dec 12, 2022



Abstract:Prostate cancer (PCa) is one of the most prevalent cancers in men and many people around the world die from clinically significant PCa (csPCa). Early diagnosis of csPCa in bi-parametric MRI (bpMRI), which is non-invasive, cost-effective, and more efficient compared to multiparametric MRI (mpMRI), can contribute to precision care for PCa. The rapid rise in artificial intelligence (AI) algorithms are enabling unprecedented improvements in providing decision support systems that can aid in csPCa diagnosis and understanding. However, existing state of the art AI algorithms which are based on deep learning technology are often limited to 2D images that fails to capture inter-slice correlations in 3D volumetric images. The use of 3D convolutional neural networks (CNNs) partly overcomes this limitation, but it does not adapt to the anisotropy of images, resulting in sub-optimal semantic representation and poor generalization. Furthermore, due to the limitation of the amount of labelled data of bpMRI and the difficulty of labelling, existing CNNs are built on relatively small datasets, leading to a poor performance. To address the limitations identified above, we propose a new Zonal-aware Self-supervised Mesh Network (Z-SSMNet) that adaptatively fuses multiple 2D, 2.5D and 3D CNNs to effectively balance representation for sparse inter-slice information and dense intra-slice information in bpMRI. A self-supervised learning (SSL) technique is further introduced to pre-train our network using unlabelled data to learn the generalizable image features. Furthermore, we constrained our network to understand the zonal specific domain knowledge to improve the diagnosis precision of csPCa. Experiments on the PI-CAI Challenge dataset demonstrate our proposed method achieves better performance for csPCa detection and diagnosis in bpMRI.
Unsupervised Representation Learning for 3D MRI Super Resolution with Degradation Adaptation
May 13, 2022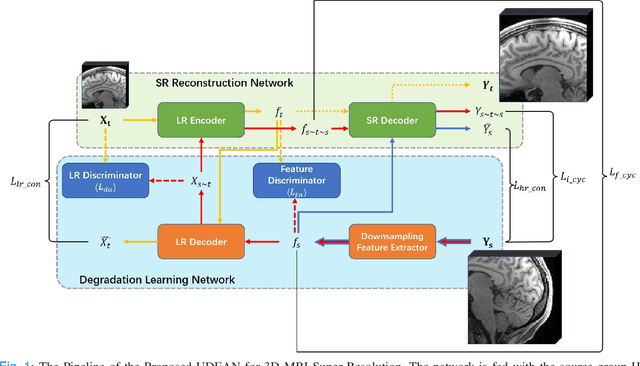
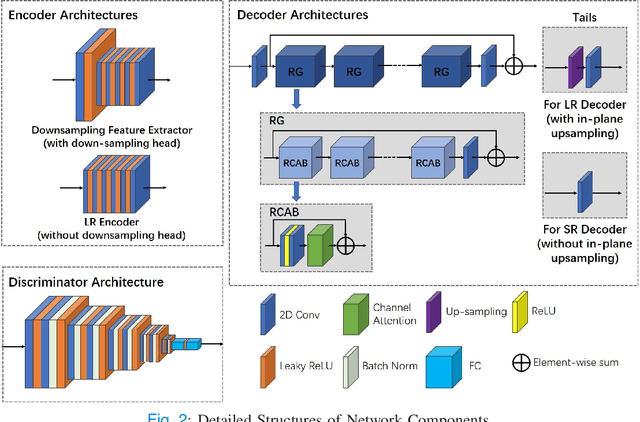
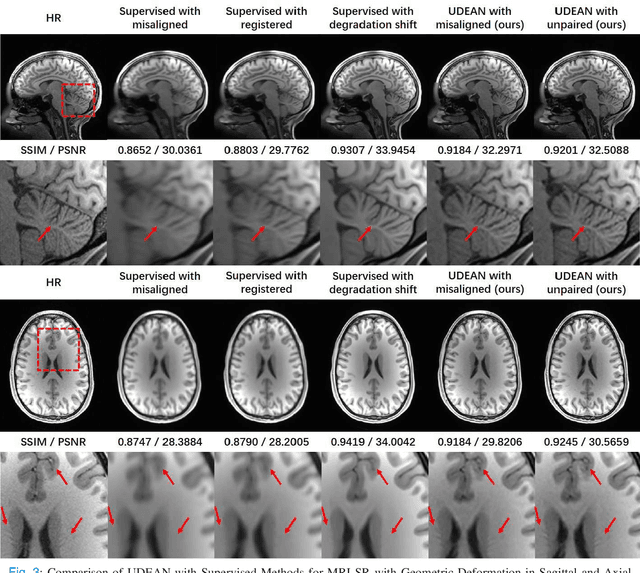
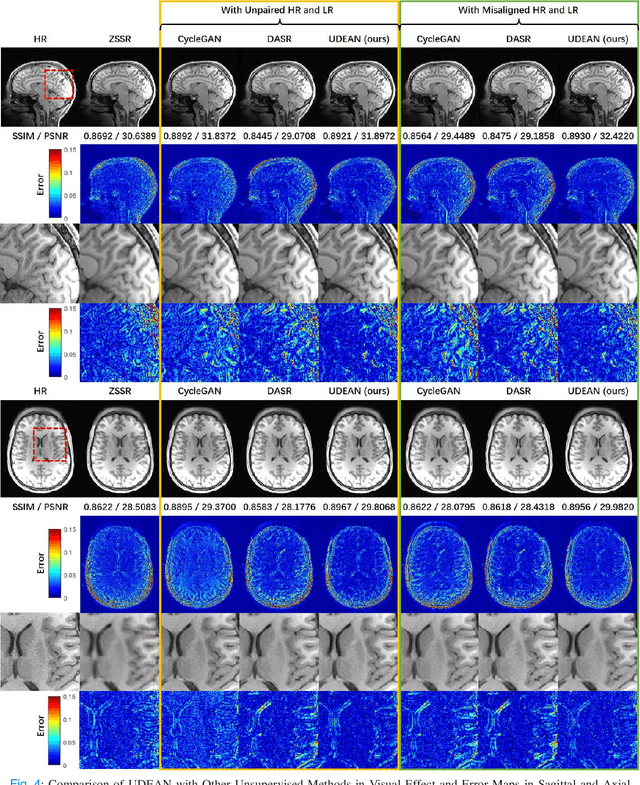
Abstract:High-resolution (HR) MRI is critical in assisting the doctor's diagnosis and image-guided treatment, but is hard to obtain in a clinical setting due to long acquisition time. Therefore, the research community investigated deep learning-based super-resolution (SR) technology to reconstruct HR MRI images with shortened acquisition time. However, training such neural networks usually requires paired HR and low-resolution (LR) in-vivo images, which are difficult to acquire due to patient movement during and between the image acquisition. Rigid movements of hard tissues can be corrected with image-registration, whereas the alignment of deformed soft tissues is challenging, making it impractical to train the neural network with such authentic HR and LR image pairs. Therefore, most of the previous studies proposed SR reconstruction by employing authentic HR images and synthetic LR images downsampled from the HR images, yet the difference in degradation representations between synthetic and authentic LR images suppresses the performance of SR reconstruction from authentic LR images. To mitigate the aforementioned problems, we propose a novel Unsupervised DEgradation Adaptation Network (UDEAN). Our model consists of two components: the degradation learning network and the SR reconstruction network. The degradation learning network downsamples the HR images by addressing the degradation representation of the misaligned or unpaired LR images, and the SR reconstruction network learns the mapping from the downsampled HR images to their original HR images. As a result, the SR reconstruction network can generate SR images from the LR images and achieve comparable quality to the HR images. Experimental results show that our method outperforms the state-of-the-art models and can potentially be applied in real-world clinical settings.
Self-Supervised Learning Framework for Remote Heart Rate Estimation Using Spatiotemporal Augmentation
Jul 16, 2021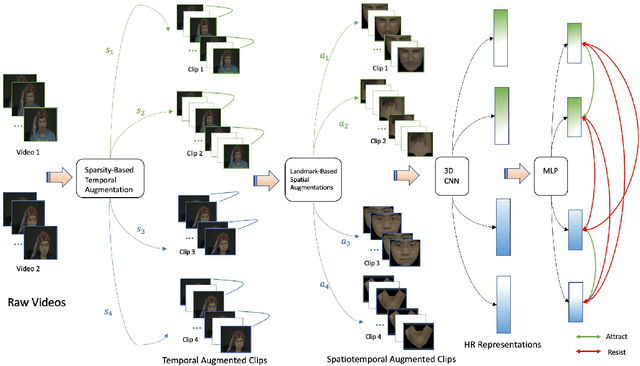



Abstract:Recent supervised deep learning methods have shown that heart rate can be measured remotely using facial videos. However, the performance of these supervised method are dependent on the availability of large-scale labelled data and they have been limited to 2D deep learning architectures that do not fully exploit the 3D spatiotemporal information. To solve this problem, we present a novel 3D self-supervised spatiotemporal learning framework for remote HR estimation on facial videos. Concretely, we propose a landmark-based spatial augmentation which splits the face into several informative parts based on the Shafer's dichromatic reflection model and a novel sparsity-based temporal augmentation exploiting Nyquist-Shannon sampling theorem to enhance the signal modelling ability. We evaluated our method on 3 public datasets and outperformed other self-supervised methods and achieved competitive accuracy with the state-of-the-art supervised methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge