Shuchang Ye
Beyond Calibration: Confounding Pathology Limits Foundation Model Specificity in Abdominal Trauma CT
Feb 10, 2026Abstract:Purpose: Translating foundation models into clinical practice requires evaluating their performance under compound distribution shift, where severe class imbalance coexists with heterogeneous imaging appearances. This challenge is relevant for traumatic bowel injury, a rare but high-mortality diagnosis. We investigated whether specificity deficits in foundation models are associated with heterogeneity in the negative class. Methods: This retrospective study used the multi-institutional, RSNA Abdominal Traumatic Injury CT dataset (2019-2023), comprising scans from 23 centres. Two foundation models (MedCLIP, zero-shot; RadDINO, linear probe) were compared against three task-specific approaches (CNN, Transformer, Ensemble). Models were trained on 3,147 patients (2.3% bowel injury prevalence) and evaluated on an enriched 100-patient test set. To isolate negative-class effects, specificity was assessed in patients without bowel injury who had concurrent solid organ injury (n=58) versus no abdominal pathology (n=50). Results: Foundation models achieved equivalent discrimination to task-specific models (AUC, 0.64-0.68 versus 0.58-0.64) with higher sensitivity (79-91% vs 41-74%) but lower specificity (33-50% vs 50-88%). All models demonstrated high specificity in patients without abdominal pathology (84-100%). When solid organ injuries were present, specificity declined substantially for foundation models (50-51 percentage points) compared with smaller reductions of 12-41 percentage points for task-specific models. Conclusion: Foundation models matched task-specific discrimination without task-specific training, but their specificity deficits were driven primarily by confounding negative-class heterogeneity rather than prevalence alone. Susceptibility to negative-class heterogeneity decreased progressively with labelled training, suggesting adaptation is required before clinical implementation.
MRG-R1: Reinforcement Learning for Clinically Aligned Medical Report Generation
Dec 18, 2025Abstract:Medical report generation (MRG) aims to automatically derive radiology-style reports from medical images to aid in clinical decision-making. However, existing methods often generate text that mimics the linguistic style of radiologists but fails to guarantee clinical correctness, because they are trained on token-level objectives which focus on word-choice and sentence structure rather than actual medical accuracy. We propose a semantic-driven reinforcement learning (SRL) method for medical report generation, adopted on a large vision-language model (LVLM). SRL adopts Group Relative Policy Optimization (GRPO) to encourage clinical-correctness-guided learning beyond imitation of language style. Specifically, we optimise a report-level reward: a margin-based cosine similarity (MCCS) computed between key radiological findings extracted from generated and reference reports, thereby directly aligning clinical-label agreement and improving semantic correctness. A lightweight reasoning format constraint further guides the model to generate structured "thinking report" outputs. We evaluate Medical Report Generation with Sematic-driven Reinforment Learning (MRG-R1), on two datasets: IU X-Ray and MIMIC-CXR using clinical efficacy (CE) metrics. MRG-R1 achieves state-of-the-art performance with CE-F1 51.88 on IU X-Ray and 40.39 on MIMIC-CXR. We found that the label-semantic reinforcement is better than conventional token-level supervision. These results indicate that optimizing a clinically grounded, report-level reward rather than token overlap,meaningfully improves clinical correctness. This work is a prior to explore semantic-reinforcement in supervising medical correctness in medical Large vision-language model(Med-LVLM) training.
MRGAgents: A Multi-Agent Framework for Improved Medical Report Generation with Med-LVLMs
May 24, 2025Abstract:Medical Large Vision-Language Models (Med-LVLMs) have been widely adopted for medical report generation. Despite Med-LVLMs producing state-of-the-art performance, they exhibit a bias toward predicting all findings as normal, leading to reports that overlook critical abnormalities. Furthermore, these models often fail to provide comprehensive descriptions of radiologically relevant regions necessary for accurate diagnosis. To address these challenges, we proposeMedical Report Generation Agents (MRGAgents), a novel multi-agent framework that fine-tunes specialized agents for different disease categories. By curating subsets of the IU X-ray and MIMIC-CXR datasets to train disease-specific agents, MRGAgents generates reports that more effectively balance normal and abnormal findings while ensuring a comprehensive description of clinically relevant regions. Our experiments demonstrate that MRGAgents outperformed the state-of-the-art, improving both report comprehensiveness and diagnostic utility.
MedCFVQA: A Causal Approach to Mitigate Modality Preference Bias in Medical Visual Question Answering
May 23, 2025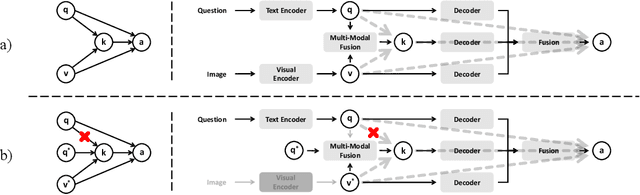
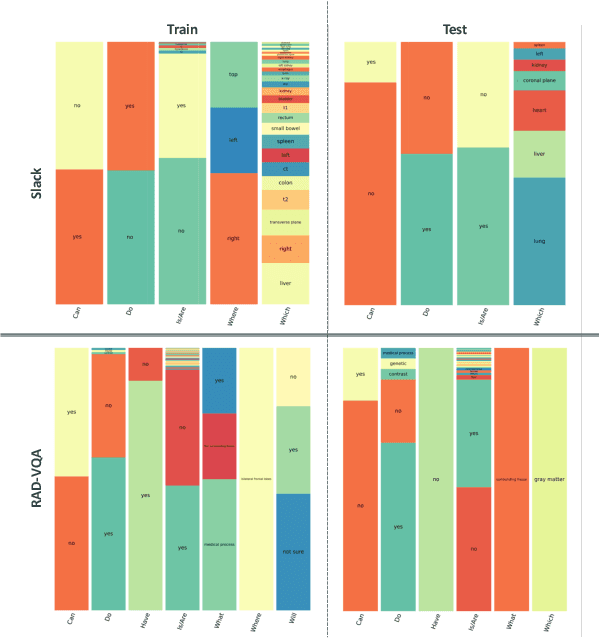
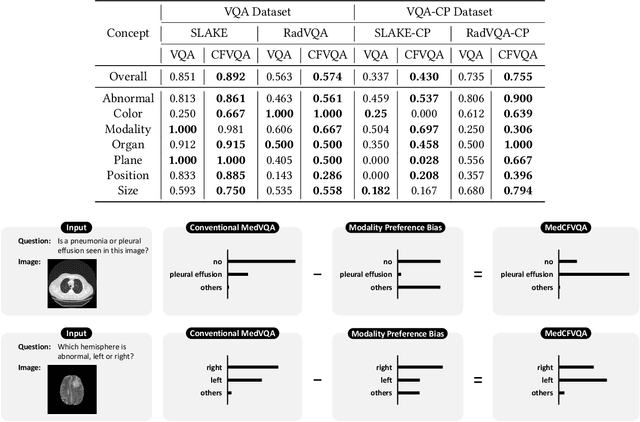
Abstract:Medical Visual Question Answering (MedVQA) is crucial for enhancing the efficiency of clinical diagnosis by providing accurate and timely responses to clinicians' inquiries regarding medical images. Existing MedVQA models suffered from modality preference bias, where predictions are heavily dominated by one modality while overlooking the other (in MedVQA, usually questions dominate the answer but images are overlooked), thereby failing to learn multimodal knowledge. To overcome the modality preference bias, we proposed a Medical CounterFactual VQA (MedCFVQA) model, which trains with bias and leverages causal graphs to eliminate the modality preference bias during inference. Existing MedVQA datasets exhibit substantial prior dependencies between questions and answers, which results in acceptable performance even if the model significantly suffers from the modality preference bias. To address this issue, we reconstructed new datasets by leveraging existing MedVQA datasets and Changed their P3rior dependencies (CP) between questions and their answers in the training and test set. Extensive experiments demonstrate that MedCFVQA significantly outperforms its non-causal counterpart on both SLAKE, RadVQA and SLAKE-CP, RadVQA-CP datasets.
A Causal Approach to Mitigate Modality Preference Bias in Medical Visual Question Answering
May 22, 2025


Abstract:Medical Visual Question Answering (MedVQA) is crucial for enhancing the efficiency of clinical diagnosis by providing accurate and timely responses to clinicians' inquiries regarding medical images. Existing MedVQA models suffered from modality preference bias, where predictions are heavily dominated by one modality while overlooking the other (in MedVQA, usually questions dominate the answer but images are overlooked), thereby failing to learn multimodal knowledge. To overcome the modality preference bias, we proposed a Medical CounterFactual VQA (MedCFVQA) model, which trains with bias and leverages causal graphs to eliminate the modality preference bias during inference. Existing MedVQA datasets exhibit substantial prior dependencies between questions and answers, which results in acceptable performance even if the model significantly suffers from the modality preference bias. To address this issue, we reconstructed new datasets by leveraging existing MedVQA datasets and Changed their P3rior dependencies (CP) between questions and their answers in the training and test set. Extensive experiments demonstrate that MedCFVQA significantly outperforms its non-causal counterpart on both SLAKE, RadVQA and SLAKE-CP, RadVQA-CP datasets.
MAISY: Motion-Aware Image SYnthesis for Medical Image Motion Correction
May 08, 2025Abstract:Patient motion during medical image acquisition causes blurring, ghosting, and distorts organs, which makes image interpretation challenging. Current state-of-the-art algorithms using Generative Adversarial Network (GAN)-based methods with their ability to learn the mappings between corrupted images and their ground truth via Structural Similarity Index Measure (SSIM) loss effectively generate motion-free images. However, we identified the following limitations: (i) they mainly focus on global structural characteristics and therefore overlook localized features that often carry critical pathological information, and (ii) the SSIM loss function struggles to handle images with varying pixel intensities, luminance factors, and variance. In this study, we propose Motion-Aware Image SYnthesis (MAISY) which initially characterize motion and then uses it for correction by: (a) leveraging the foundation model Segment Anything Model (SAM), to dynamically learn spatial patterns along anatomical boundaries where motion artifacts are most pronounced and, (b) introducing the Variance-Selective SSIM (VS-SSIM) loss which adaptively emphasizes spatial regions with high pixel variance to preserve essential anatomical details during artifact correction. Experiments on chest and head CT datasets demonstrate that our model outperformed the state-of-the-art counterparts, with Peak Signal-to-Noise Ratio (PSNR) increasing by 40%, SSIM by 10%, and Dice by 16%.
SMPL-GPTexture: Dual-View 3D Human Texture Estimation using Text-to-Image Generation Models
Apr 17, 2025Abstract:Generating high-quality, photorealistic textures for 3D human avatars remains a fundamental yet challenging task in computer vision and multimedia field. However, real paired front and back images of human subjects are rarely available with privacy, ethical and cost of acquisition, which restricts scalability of the data. Additionally, learning priors from image inputs using deep generative models, such as GANs or diffusion models, to infer unseen regions such as the human back often leads to artifacts, structural inconsistencies, or loss of fine-grained detail. To address these issues, we present SMPL-GPTexture (skinned multi-person linear model - general purpose Texture), a novel pipeline that takes natural language prompts as input and leverages a state-of-the-art text-to-image generation model to produce paired high-resolution front and back images of a human subject as the starting point for texture estimation. Using the generated paired dual-view images, we first employ a human mesh recovery model to obtain a robust 2D-to-3D SMPL alignment between image pixels and the 3D model's UV coordinates for each views. Second, we use an inverted rasterization technique that explicitly projects the observed colour from the input images into the UV space, thereby producing accurate, complete texture maps. Finally, we apply a diffusion-based inpainting module to fill in the missing regions, and the fusion mechanism then combines these results into a unified full texture map. Extensive experiments shows that our SMPL-GPTexture can generate high resolution texture aligned with user's prompts.
Language-guided Medical Image Segmentation with Target-informed Multi-level Contrastive Alignments
Dec 18, 2024



Abstract:Medical image segmentation is crucial in modern medical image analysis, which can aid into diagnosis of various disease conditions. Recently, language-guided segmentation methods have shown promising results in automating image segmentation where text reports are incorporated as guidance. These text reports, containing image impressions and insights given by clinicians, provides auxiliary guidance. However, these methods neglect the inherent pattern gaps between the two distinct modalities, which leads to sub-optimal image-text feature fusion without proper cross-modality feature alignments. Contrastive alignments are widely used to associate image-text semantics in representation learning; however, it has not been exploited to bridge the pattern gaps in language-guided segmentation that relies on subtle low level image details to represent diseases. Existing contrastive alignment methods typically algin high-level global image semantics without involving low-level, localized target information, and therefore fails to explore fine-grained text guidance for language-guided segmentation. In this study, we propose a language-guided segmentation network with Target-informed Multi-level Contrastive Alignments (TMCA). TMCA enables target-informed cross-modality alignments and fine-grained text guidance to bridge the pattern gaps in language-guided segmentation. Specifically, we introduce: 1) a target-sensitive semantic distance module that enables granular image-text alignment modelling, and 2) a multi-level alignment strategy that directs text guidance on low-level image features. In addition, a language-guided target enhancement module is proposed to leverage the aligned text to redirect attention to focus on critical localized image features. Extensive experiments on 4 image-text datasets, involving 3 medical imaging modalities, demonstrated that our TMCA achieved superior performances.
SGSeg: Enabling Text-free Inference in Language-guided Segmentation of Chest X-rays via Self-guidance
Sep 07, 2024Abstract:Segmentation of infected areas in chest X-rays is pivotal for facilitating the accurate delineation of pulmonary structures and pathological anomalies. Recently, multi-modal language-guided image segmentation methods have emerged as a promising solution for chest X-rays where the clinical text reports, depicting the assessment of the images, are used as guidance. Nevertheless, existing language-guided methods require clinical reports alongside the images, and hence, they are not applicable for use in image segmentation in a decision support context, but rather limited to retrospective image analysis after clinical reporting has been completed. In this study, we propose a self-guided segmentation framework (SGSeg) that leverages language guidance for training (multi-modal) while enabling text-free inference (uni-modal), which is the first that enables text-free inference in language-guided segmentation. We exploit the critical location information of both pulmonary and pathological structures depicted in the text reports and introduce a novel localization-enhanced report generation (LERG) module to generate clinical reports for self-guidance. Our LERG integrates an object detector and a location-based attention aggregator, weakly-supervised by a location-aware pseudo-label extraction module. Extensive experiments on a well-benchmarked QaTa-COV19 dataset demonstrate that our SGSeg achieved superior performance than existing uni-modal segmentation methods and closely matched the state-of-the-art performance of multi-modal language-guided segmentation methods.
Dual-modal Dynamic Traceback Learning for Medical Report Generation
Jan 24, 2024Abstract:With increasing reliance on medical imaging in clinical practices, automated report generation from medical images is in great demand. Existing report generation methods typically adopt an encoder-decoder deep learning framework to build a uni-directional image-to-report mapping. However, such a framework ignores the bi-directional mutual associations between images and reports, thus incurring difficulties in associating the intrinsic medical meanings between them. Recent generative representation learning methods have demonstrated the benefits of dual-modal learning from both image and text modalities. However, these methods exhibit two major drawbacks for medical report generation: 1) they tend to capture morphological information and have difficulties in capturing subtle pathological semantic information, and 2) they predict masked text rely on both unmasked images and text, inevitably degrading performance when inference is based solely on images. In this study, we propose a new report generation framework with dual-modal dynamic traceback learning (DTrace) to overcome the two identified drawbacks and enable dual-modal learning for medical report generation. To achieve this, our DTrace introduces a traceback mechanism to control the semantic validity of generated content via self-assessment. Further, our DTrace introduces a dynamic learning strategy to adapt to various proportions of image and text input, enabling report generation without reliance on textual input during inference. Extensive experiments on two well-benchmarked datasets (IU-Xray and MIMIC-CXR) show that our DTrace outperforms state-of-the-art medical report generation methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge