David Dagan Feng
Language-guided Medical Image Segmentation with Target-informed Multi-level Contrastive Alignments
Dec 18, 2024



Abstract:Medical image segmentation is crucial in modern medical image analysis, which can aid into diagnosis of various disease conditions. Recently, language-guided segmentation methods have shown promising results in automating image segmentation where text reports are incorporated as guidance. These text reports, containing image impressions and insights given by clinicians, provides auxiliary guidance. However, these methods neglect the inherent pattern gaps between the two distinct modalities, which leads to sub-optimal image-text feature fusion without proper cross-modality feature alignments. Contrastive alignments are widely used to associate image-text semantics in representation learning; however, it has not been exploited to bridge the pattern gaps in language-guided segmentation that relies on subtle low level image details to represent diseases. Existing contrastive alignment methods typically algin high-level global image semantics without involving low-level, localized target information, and therefore fails to explore fine-grained text guidance for language-guided segmentation. In this study, we propose a language-guided segmentation network with Target-informed Multi-level Contrastive Alignments (TMCA). TMCA enables target-informed cross-modality alignments and fine-grained text guidance to bridge the pattern gaps in language-guided segmentation. Specifically, we introduce: 1) a target-sensitive semantic distance module that enables granular image-text alignment modelling, and 2) a multi-level alignment strategy that directs text guidance on low-level image features. In addition, a language-guided target enhancement module is proposed to leverage the aligned text to redirect attention to focus on critical localized image features. Extensive experiments on 4 image-text datasets, involving 3 medical imaging modalities, demonstrated that our TMCA achieved superior performances.
Automatic Left Ventricular Cavity Segmentation via Deep Spatial Sequential Network in 4D Computed Tomography Studies
Dec 17, 2024



Abstract:Automated segmentation of left ventricular cavity (LVC) in temporal cardiac image sequences (multiple time points) is a fundamental requirement for quantitative analysis of its structural and functional changes. Deep learning based methods for the segmentation of LVC are the state of the art; however, these methods are generally formulated to work on single time points, and fails to exploit the complementary information from the temporal image sequences that can aid in segmentation accuracy and consistency among the images across the time points. Furthermore, these segmentation methods perform poorly in segmenting the end-systole (ES) phase images, where the left ventricle deforms to the smallest irregular shape, and the boundary between the blood chamber and myocardium becomes inconspicuous. To overcome these limitations, we propose a new method to automatically segment temporal cardiac images where we introduce a spatial sequential (SS) network to learn the deformation and motion characteristics of the LVC in an unsupervised manner; these characteristics were then integrated with sequential context information derived from bi-directional learning (BL) where both chronological and reverse-chronological directions of the image sequence were used. Our experimental results on a cardiac computed tomography (CT) dataset demonstrated that our spatial-sequential network with bi-directional learning (SS-BL) method outperformed existing methods for LVC segmentation. Our method was also applied to MRI cardiac dataset and the results demonstrated the generalizability of our method.
Enhancing medical vision-language contrastive learning via inter-matching relation modelling
Jan 19, 2024Abstract:Medical image representations can be learned through medical vision-language contrastive learning (mVLCL) where medical imaging reports are used as weak supervision through image-text alignment. These learned image representations can be transferred to and benefit various downstream medical vision tasks such as disease classification and segmentation. Recent mVLCL methods attempt to align image sub-regions and the report keywords as local-matchings. However, these methods aggregate all local-matchings via simple pooling operations while ignoring the inherent relations between them. These methods therefore fail to reason between local-matchings that are semantically related, e.g., local-matchings that correspond to the disease word and the location word (semantic-relations), and also fail to differentiate such clinically important local-matchings from others that correspond to less meaningful words, e.g., conjunction words (importance-relations). Hence, we propose a mVLCL method that models the inter-matching relations between local-matchings via a relation-enhanced contrastive learning framework (RECLF). In RECLF, we introduce a semantic-relation reasoning module (SRM) and an importance-relation reasoning module (IRM) to enable more fine-grained report supervision for image representation learning. We evaluated our method using four public benchmark datasets on four downstream tasks, including segmentation, zero-shot classification, supervised classification, and cross-modal retrieval. Our results demonstrated the superiority of our RECLF over the state-of-the-art mVLCL methods with consistent improvements across single-modal and cross-modal tasks. These results suggest that our RECLF, by modelling the inter-matching relations, can learn improved medical image representations with better generalization capabilities.
PET Synthesis via Self-supervised Adaptive Residual Estimation Generative Adversarial Network
Oct 24, 2023



Abstract:Positron emission tomography (PET) is a widely used, highly sensitive molecular imaging in clinical diagnosis. There is interest in reducing the radiation exposure from PET but also maintaining adequate image quality. Recent methods using convolutional neural networks (CNNs) to generate synthesized high-quality PET images from low-dose counterparts have been reported to be state-of-the-art for low-to-high image recovery methods. However, these methods are prone to exhibiting discrepancies in texture and structure between synthesized and real images. Furthermore, the distribution shift between low-dose PET and standard PET has not been fully investigated. To address these issues, we developed a self-supervised adaptive residual estimation generative adversarial network (SS-AEGAN). We introduce (1) An adaptive residual estimation mapping mechanism, AE-Net, designed to dynamically rectify the preliminary synthesized PET images by taking the residual map between the low-dose PET and synthesized output as the input, and (2) A self-supervised pre-training strategy to enhance the feature representation of the coarse generator. Our experiments with a public benchmark dataset of total-body PET images show that SS-AEGAN consistently outperformed the state-of-the-art synthesis methods with various dose reduction factors.
Deep Dynamic Epidemiological Modelling for COVID-19 Forecasting in Multi-level Districts
Jun 21, 2023



Abstract:Objective: COVID-19 has spread worldwide and made a huge influence across the world. Modeling the infectious spread situation of COVID-19 is essential to understand the current condition and to formulate intervention measurements. Epidemiological equations based on the SEIR model simulate disease development. The traditional parameter estimation method to solve SEIR equations could not precisely fit real-world data due to different situations, such as social distancing policies and intervention strategies. Additionally, learning-based models achieve outstanding fitting performance, but cannot visualize mechanisms. Methods: Thus, we propose a deep dynamic epidemiological (DDE) method that combines epidemiological equations and deep-learning advantages to obtain high accuracy and visualization. The DDE contains deep networks to fit the effect function to simulate the ever-changing situations based on the neural ODE method in solving variants' equations, ensuring the fitting performance of multi-level areas. Results: We introduce four SEIR variants to fit different situations in different countries and regions. We compare our DDE method with traditional parameter estimation methods (Nelder-Mead, BFGS, Powell, Truncated Newton Conjugate-Gradient, Neural ODE) in fitting the real-world data in the cases of countries (the USA, Columbia, South Africa) and regions (Wuhan in China, Piedmont in Italy). Our DDE method achieves the best Mean Square Error and Pearson coefficient in all five areas. Further, compared with the state-of-art learning-based approaches, the DDE outperforms all techniques, including LSTM, RNN, GRU, Random Forest, Extremely Random Trees, and Decision Tree. Conclusion: DDE presents outstanding predictive ability and visualized display of the changes in infection rates in different regions and countries.
Hyper-Connected Transformer Network for Co-Learning Multi-Modality PET-CT Features
Oct 28, 2022



Abstract:[18F]-Fluorodeoxyglucose (FDG) positron emission tomography - computed tomography (PET-CT) has become the imaging modality of choice for diagnosing many cancers. Co-learning complementary PET-CT imaging features is a fundamental requirement for automatic tumor segmentation and for developing computer aided cancer diagnosis systems. We propose a hyper-connected transformer (HCT) network that integrates a transformer network (TN) with a hyper connected fusion for multi-modality PET-CT images. The TN was leveraged for its ability to provide global dependencies in image feature learning, which was achieved by using image patch embeddings with a self-attention mechanism to capture image-wide contextual information. We extended the single-modality definition of TN with multiple TN based branches to separately extract image features. We introduced a hyper connected fusion to fuse the contextual and complementary image features across multiple transformers in an iterative manner. Our results with two non-small cell lung cancer and soft-tissue sarcoma datasets show that HCT achieved better performance in segmentation accuracy when compared to state-of-the-art methods. We also show that HCT produces consistent performance across various image fusion strategies and network backbones.
DeepMTS: Deep Multi-task Learning for Survival Prediction in Patients with Advanced Nasopharyngeal Carcinoma using Pretreatment PET/CT
Sep 16, 2021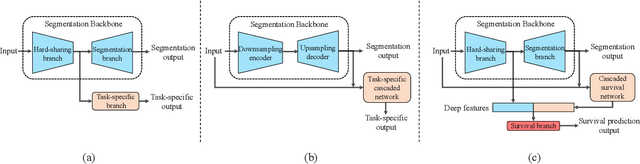
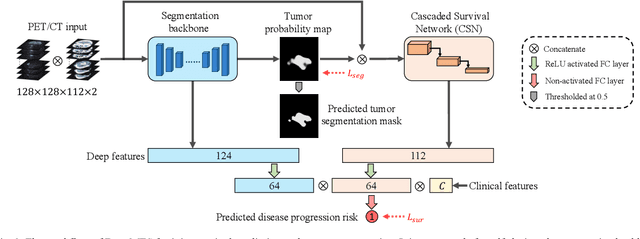
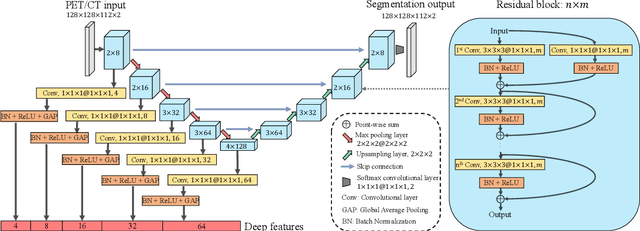
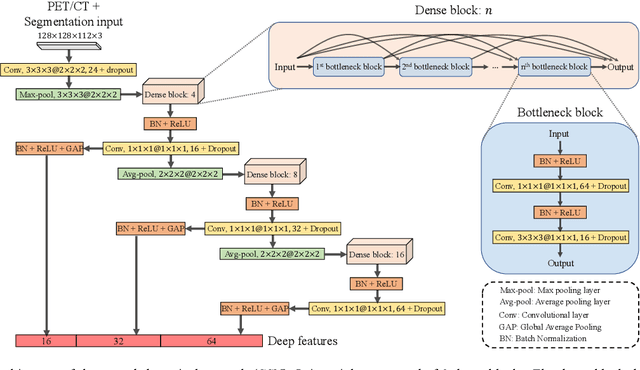
Abstract:Nasopharyngeal Carcinoma (NPC) is a worldwide malignant epithelial cancer. Survival prediction is a major concern for NPC patients, as it provides early prognostic information that is needed to guide treatments. Recently, deep learning, which leverages Deep Neural Networks (DNNs) to learn deep representations of image patterns, has been introduced to the survival prediction in various cancers including NPC. It has been reported that image-derived end-to-end deep survival models have the potential to outperform clinical prognostic indicators and traditional radiomics-based survival models in prognostic performance. However, deep survival models, especially 3D models, require large image training data to avoid overfitting. Unfortunately, medical image data is usually scarce, especially for Positron Emission Tomography/Computed Tomography (PET/CT) due to the high cost of PET/CT scanning. Compared to Magnetic Resonance Imaging (MRI) or Computed Tomography (CT) providing only anatomical information of tumors, PET/CT that provides both anatomical (from CT) and metabolic (from PET) information is promising to achieve more accurate survival prediction. However, we have not identified any 3D end-to-end deep survival model that applies to small PET/CT data of NPC patients. In this study, we introduced the concept of multi-task leaning into deep survival models to address the overfitting problem resulted from small data. Tumor segmentation was incorporated as an auxiliary task to enhance the model's efficiency of learning from scarce PET/CT data. Based on this idea, we proposed a 3D end-to-end Deep Multi-Task Survival model (DeepMTS) for joint survival prediction and tumor segmentation. Our DeepMTS can jointly learn survival prediction and tumor segmentation using PET/CT data of only 170 patients with advanced NPC.
Prediction of 5-year Progression-Free Survival in Advanced Nasopharyngeal Carcinoma with Pretreatment PET/CT using Multi-Modality Deep Learning-based Radiomics
Mar 09, 2021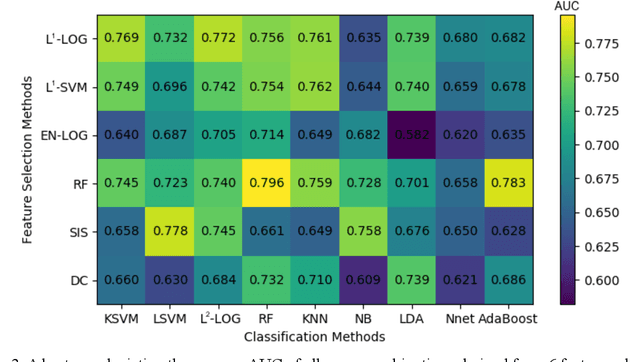
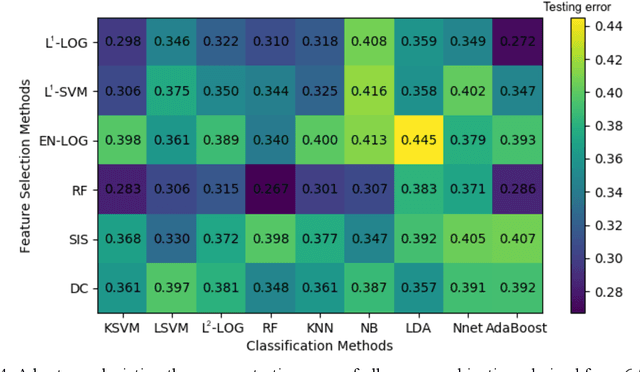
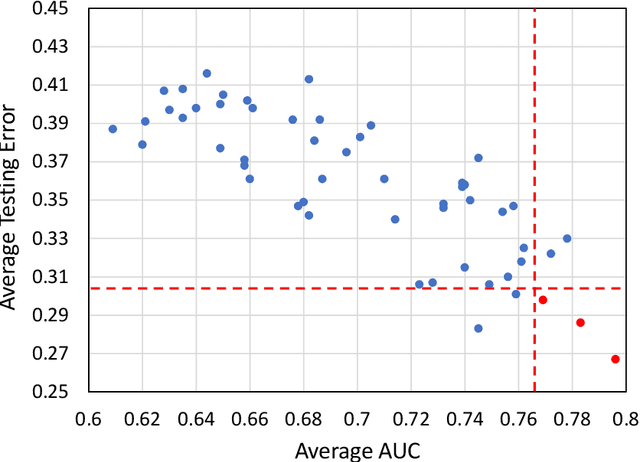
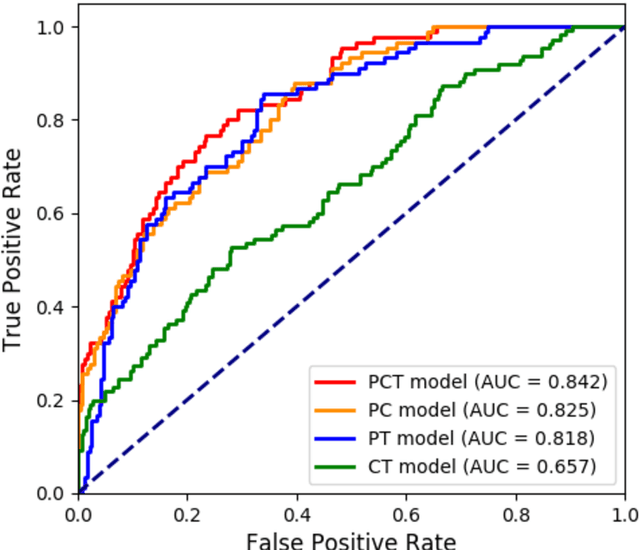
Abstract:Deep Learning-based Radiomics (DLR) has achieved great success on medical image analysis. In this study, we aim to explore the capability of DLR for survival prediction in NPC. We developed an end-to-end multi-modality DLR model using pretreatment PET/CT images to predict 5-year Progression-Free Survival (PFS) in advanced NPC. A total of 170 patients with pathological confirmed advanced NPC (TNM stage III or IVa) were enrolled in this study. A 3D Convolutional Neural Network (CNN), with two branches to process PET and CT separately, was optimized to extract deep features from pretreatment multi-modality PET/CT images and use the derived features to predict the probability of 5-year PFS. Optionally, TNM stage, as a high-level clinical feature, can be integrated into our DLR model to further improve prognostic performance. For a comparison between CR and DLR, 1456 handcrafted features were extracted, and three top CR methods were selected as benchmarks from 54 combinations of 6 feature selection methods and 9 classification methods. Compared to the three CR methods, our multi-modality DLR models using both PET and CT, with or without TNM stage (named PCT or PC model), resulted in the highest prognostic performance. Furthermore, the multi-modality PCT model outperformed single-modality DLR models using only PET and TNM stage (PT model) or only CT and TNM stage (CT model). Our study identified potential radiomics-based prognostic model for survival prediction in advanced NPC, and suggests that DLR could serve as a tool for aiding in cancer management.
Enhancing Medical Image Registration via Appearance Adjustment Networks
Mar 09, 2021



Abstract:Deformable image registration is fundamental for many medical image analyses. A key obstacle for accurate image registration is the variations in image appearance. Recently, deep learning-based registration methods (DLRs), using deep neural networks, have computational efficiency that is several orders of magnitude greater than traditional optimization-based registration methods (ORs). A major drawback, however, of DLRs is a disregard for the target-pair-specific optimization that is inherent in ORs and instead they rely on a globally optimized network that is trained with a set of training samples to achieve faster registration. Thus, DLRs inherently have degraded ability to adapt to appearance variations and perform poorly, compared to ORs, when image pairs (fixed/moving images) have large differences in appearance. Hence, we propose an Appearance Adjustment Network (AAN) where we leverage anatomy edges, through an anatomy-constrained loss function, to generate an anatomy-preserving appearance transformation. We designed the AAN so that it can be readily inserted into a wide range of DLRs, to reduce the appearance differences between the fixed and moving images. Our AAN and DLR's network can be trained cooperatively in an unsupervised and end-to-end manner. We evaluated our AAN with two widely used DLRs - Voxelmorph (VM) and FAst IMage registration (FAIM) - on three public 3D brain magnetic resonance (MR) image datasets - IBSR18, Mindboggle101, and LPBA40. The results show that DLRs, using the AAN, improved performance and achieved higher results than state-of-the-art ORs.
Deep Multi-Scale Resemblance Network for the Sub-class Differentiation of Adrenal Masses on Computed Tomography Images
Jul 29, 2020



Abstract:Objective: The accurate classification of mass lesions in the adrenal glands ('adrenal masses'), detected with computed tomography (CT), is important for diagnosis and patient management. Adrenal masses can be benign or malignant and the benign masses have varying prevalence. Classification methods based on convolutional neural networks (CNN) are the state-of-the-art in maximizing inter-class differences in large medical imaging training datasets. The application of CNNs, to adrenal masses is challenging due to large intra-class variations, large inter-class similarities and imbalanced training data due to the size of masses. Methods: We developed a deep multi-scale resemblance network (DMRN) to overcome these limitations and leveraged paired CNNs to evaluate the intra-class similarities. We used multi-scale feature embedding to improve the inter-class separability by iteratively combining complementary information produced at different scales of the input to create structured feature descriptors. We also augmented the training data with randomly sampled paired adrenal masses to reduce the influence of imbalanced training data. We used 229 CT scans of patients with adrenal masses. Results: Our method had the best results compared to state-of-the-art methods. Conclusion: Our DMRN sub-classified adrenal masses on CT and was superior to state-of-the-art approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge