Yige Peng
HyperWalker: Dynamic Hypergraph-Based Deep Diagnosis for Multi-Hop Clinical Modeling across EHR and X-Ray in Medical VLMs
Jan 20, 2026Abstract:Automated clinical diagnosis remains a core challenge in medical AI, which usually requires models to integrate multi-modal data and reason across complex, case-specific contexts. Although recent methods have advanced medical report generation (MRG) and visual question answering (VQA) with medical vision-language models (VLMs), these methods, however, predominantly operate under a sample-isolated inference paradigm, as such processing cases independently without access to longitudinal electronic health records (EHRs) or structurally related patient examples. This paradigm limits reasoning to image-derived information alone, which ignores external complementary medical evidence for potentially more accurate diagnosis. To overcome this limitation, we propose \textbf{HyperWalker}, a \textit{Deep Diagnosis} framework that reformulates clinical reasoning via dynamic hypergraphs and test-time training. First, we construct a dynamic hypergraph, termed \textbf{iBrochure}, to model the structural heterogeneity of EHR data and implicit high-order associations among multimodal clinical information. Within this hypergraph, a reinforcement learning agent, \textbf{Walker}, navigates to and identifies optimal diagnostic paths. To ensure comprehensive coverage of diverse clinical characteristics in test samples, we incorporate a \textit{linger mechanism}, a multi-hop orthogonal retrieval strategy that iteratively selects clinically complementary neighborhood cases reflecting distinct clinical attributes. Experiments on MRG with MIMIC and medical VQA on EHRXQA demonstrate that HyperWalker achieves state-of-the-art performance. Code is available at: https://github.com/Bean-Young/HyperWalker
PET Synthesis via Self-supervised Adaptive Residual Estimation Generative Adversarial Network
Oct 24, 2023



Abstract:Positron emission tomography (PET) is a widely used, highly sensitive molecular imaging in clinical diagnosis. There is interest in reducing the radiation exposure from PET but also maintaining adequate image quality. Recent methods using convolutional neural networks (CNNs) to generate synthesized high-quality PET images from low-dose counterparts have been reported to be state-of-the-art for low-to-high image recovery methods. However, these methods are prone to exhibiting discrepancies in texture and structure between synthesized and real images. Furthermore, the distribution shift between low-dose PET and standard PET has not been fully investigated. To address these issues, we developed a self-supervised adaptive residual estimation generative adversarial network (SS-AEGAN). We introduce (1) An adaptive residual estimation mapping mechanism, AE-Net, designed to dynamically rectify the preliminary synthesized PET images by taking the residual map between the low-dose PET and synthesized output as the input, and (2) A self-supervised pre-training strategy to enhance the feature representation of the coarse generator. Our experiments with a public benchmark dataset of total-body PET images show that SS-AEGAN consistently outperformed the state-of-the-art synthesis methods with various dose reduction factors.
CG-3DSRGAN: A classification guided 3D generative adversarial network for image quality recovery from low-dose PET images
Apr 03, 2023



Abstract:Positron emission tomography (PET) is the most sensitive molecular imaging modality routinely applied in our modern healthcare. High radioactivity caused by the injected tracer dose is a major concern in PET imaging and limits its clinical applications. However, reducing the dose leads to inadequate image quality for diagnostic practice. Motivated by the need to produce high quality images with minimum low-dose, Convolutional Neural Networks (CNNs) based methods have been developed for high quality PET synthesis from its low-dose counterparts. Previous CNNs-based studies usually directly map low-dose PET into features space without consideration of different dose reduction level. In this study, a novel approach named CG-3DSRGAN (Classification-Guided Generative Adversarial Network with Super Resolution Refinement) is presented. Specifically, a multi-tasking coarse generator, guided by a classification head, allows for a more comprehensive understanding of the noise-level features present in the low-dose data, resulting in improved image synthesis. Moreover, to recover spatial details of standard PET, an auxiliary super resolution network - Contextual-Net - is proposed as a second-stage training to narrow the gap between coarse prediction and standard PET. We compared our method to the state-of-the-art methods on whole-body PET with different dose reduction factors (DRFs). Experiments demonstrate our method can outperform others on all DRF.
Automatic Tumor Segmentation via False Positive Reduction Network for Whole-Body Multi-Modal PET/CT Images
Sep 16, 2022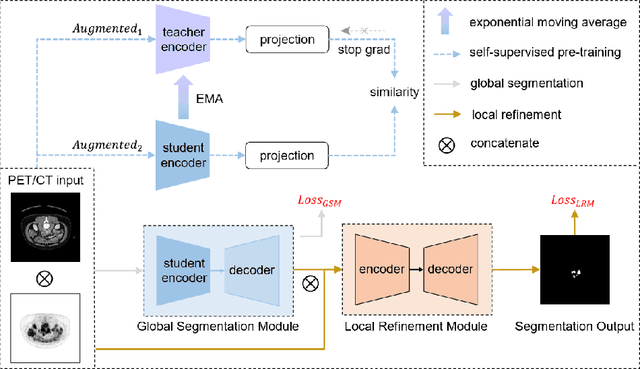
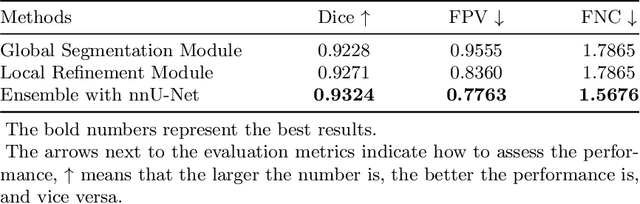
Abstract:Multi-modality Fluorodeoxyglucose (FDG) positron emission tomography / computed tomography (PET/CT) has been routinely used in the assessment of common cancers, such as lung cancer, lymphoma, and melanoma. This is mainly attributed to the fact that PET/CT combines the high sensitivity for tumor detection of PET and anatomical information from CT. In PET/CT image assessment, automatic tumor segmentation is an important step, and in recent years, deep learning based methods have become the state-of-the-art. Unfortunately, existing methods tend to over-segment the tumor regions and include regions such as the normal high uptake organs, inflammation, and other infections. In this study, we introduce a false positive reduction network to overcome this limitation. We firstly introduced a self-supervised pre-trained global segmentation module to coarsely delineate the candidate tumor regions using a self-supervised pre-trained encoder. The candidate tumor regions were then refined by removing false positives via a local refinement module. Our experiments with the MICCAI 2022 Automated Lesion Segmentation in Whole-Body FDG-PET/CT (AutoPET) challenge dataset showed that our method achieved a dice score of 0.9324 with the preliminary testing data and was ranked 1st place in dice on the leaderboard. Our method was also ranked in the top 7 methods on the final testing data, the final ranking will be announced during the 2022 MICCAI AutoPET workshop. Our code is available at: https://github.com/YigePeng/AutoPET_False_Positive_Reduction.
Predicting Distant Metastases in Soft-Tissue Sarcomas from PET-CT scans using Constrained Hierarchical Multi-Modality Feature Learning
Apr 23, 2021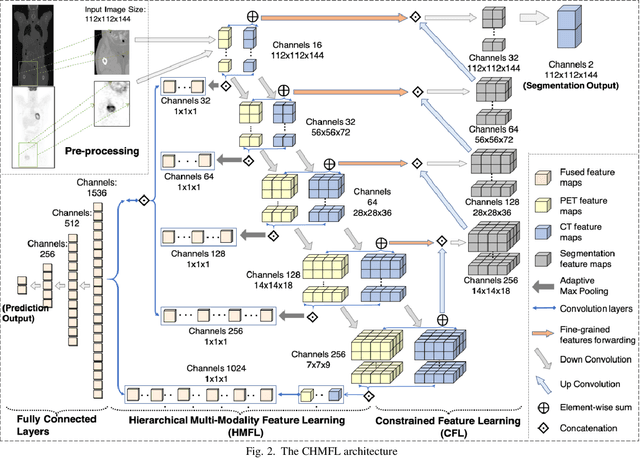
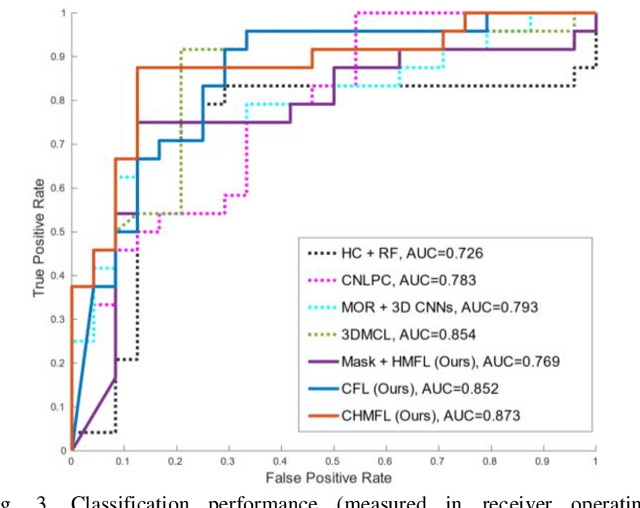
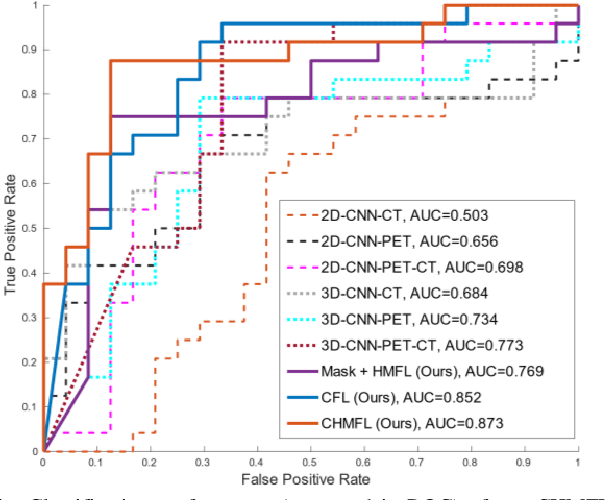
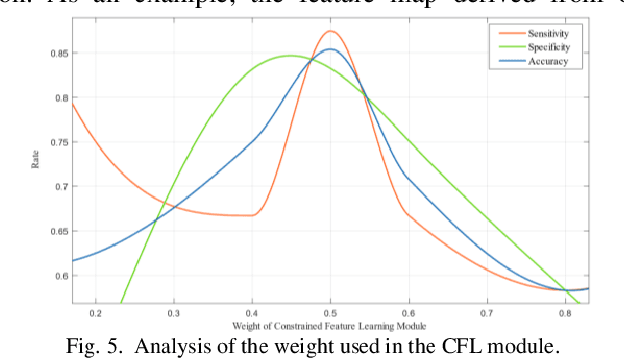
Abstract:Distant metastases (DM) refer to the dissemination of tumors, usually, beyond the organ where the tumor originated. They are the leading cause of death in patients with soft-tissue sarcomas (STSs). Positron emission tomography-computed tomography (PET-CT) is regarded as the imaging modality of choice for the management of STSs. It is difficult to determine from imaging studies which STS patients will develop metastases. 'Radiomics' refers to the extraction and analysis of quantitative features from medical images and it has been employed to help identify such tumors. The state-of-the-art in radiomics is based on convolutional neural networks (CNNs). Most CNNs are designed for single-modality imaging data (CT or PET alone) and do not exploit the information embedded in PET-CT where there is a combination of an anatomical and functional imaging modality. Furthermore, most radiomic methods rely on manual input from imaging specialists for tumor delineation, definition and selection of radiomic features. This approach, however, may not be scalable to tumors with complex boundaries and where there are multiple other sites of disease. We outline a new 3D CNN to help predict DM in STS patients from PET-CT data. The 3D CNN uses a constrained feature learning module and a hierarchical multi-modality feature learning module that leverages the complementary information from the modalities to focus on semantically important regions. Our results on a public PET-CT dataset of STS patients show that multi-modal information improves the ability to identify those patients who develop DM. Further our method outperformed all other related state-of-the-art methods.
Multi-Modality Information Fusion for Radiomics-based Neural Architecture Search
Jul 12, 2020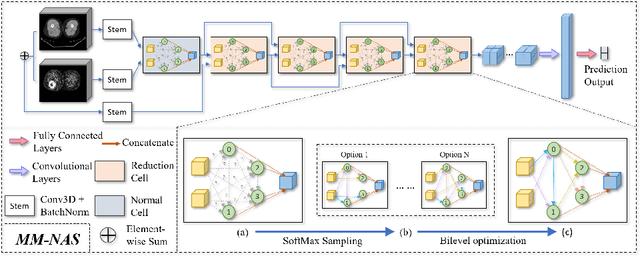
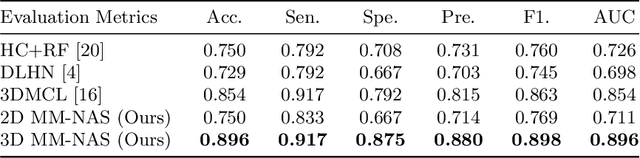
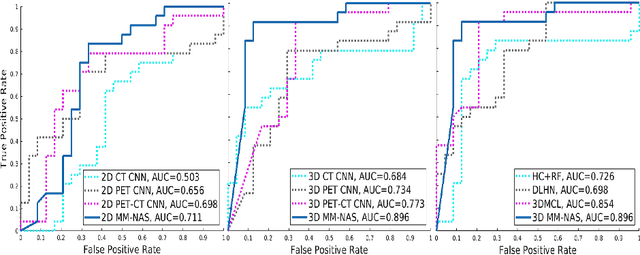
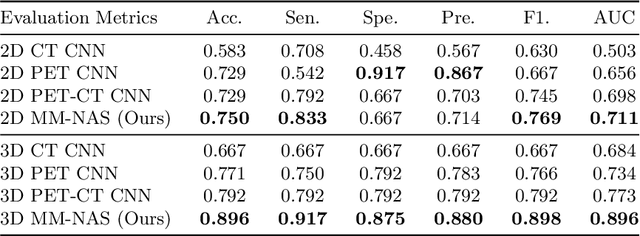
Abstract:'Radiomics' is a method that extracts mineable quantitative features from radiographic images. These features can then be used to determine prognosis, for example, predicting the development of distant metastases (DM). Existing radiomics methods, however, require complex manual effort including the design of hand-crafted radiomic features and their extraction and selection. Recent radiomics methods, based on convolutional neural networks (CNNs), also require manual input in network architecture design and hyper-parameter tuning. Radiomic complexity is further compounded when there are multiple imaging modalities, for example, combined positron emission tomography - computed tomography (PET-CT) where there is functional information from PET and complementary anatomical localization information from computed tomography (CT). Existing multi-modality radiomics methods manually fuse the data that are extracted separately. Reliance on manual fusion often results in sub-optimal fusion because they are dependent on an 'expert's' understanding of medical images. In this study, we propose a multi-modality neural architecture search method (MM-NAS) to automatically derive optimal multi-modality image features for radiomics and thus negate the dependence on a manual process. We evaluated our MM-NAS on the ability to predict DM using a public PET-CT dataset of patients with soft-tissue sarcomas (STSs). Our results show that our MM-NAS had a higher prediction accuracy when compared to state-of-the-art radiomics methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge