Ashnil Kumar
Spatio-Temporal Dual-Stream Neural Network for Sequential Whole-Body PET Segmentation
Jun 09, 2021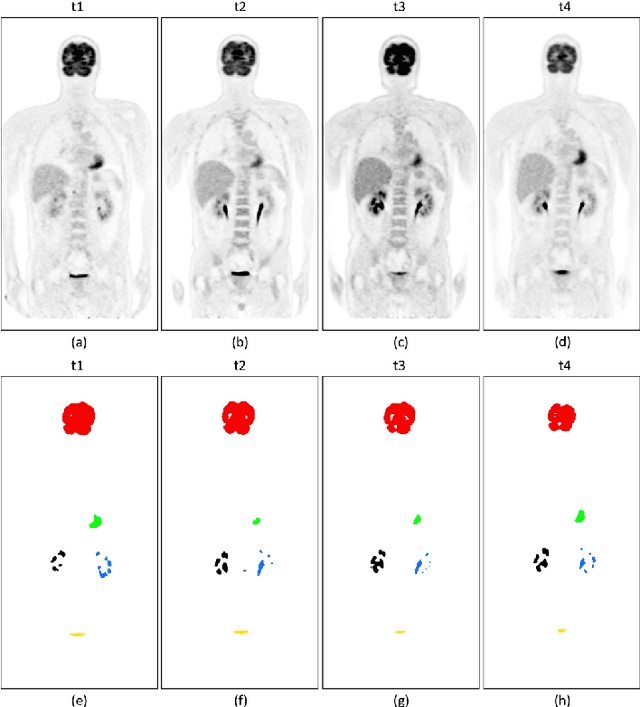
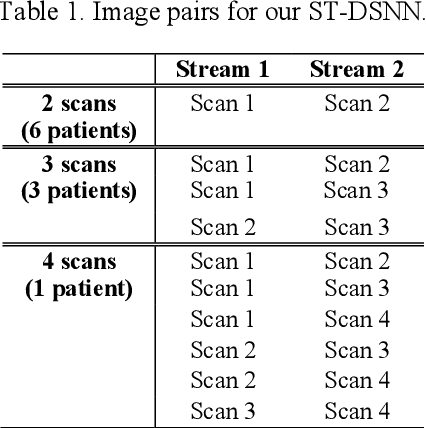
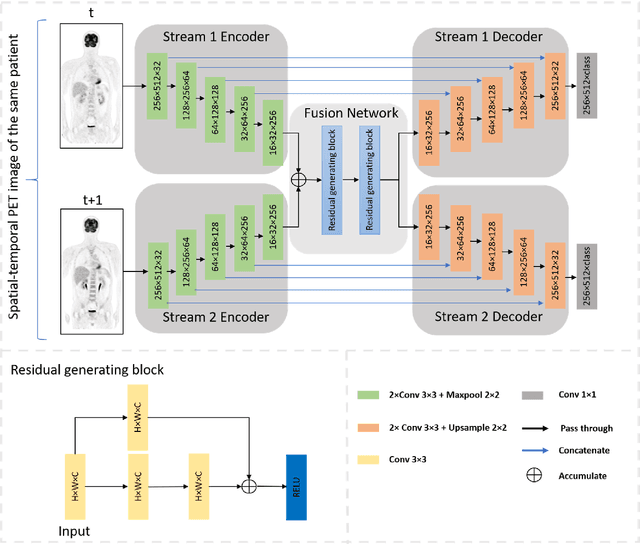
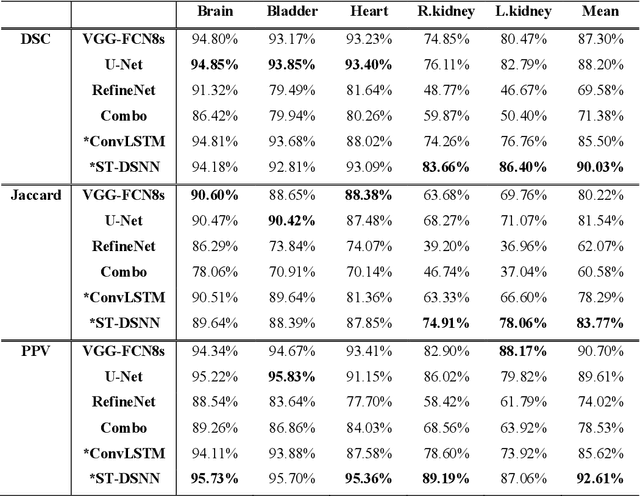
Abstract:Sequential whole-body 18F-Fluorodeoxyglucose (FDG) positron emission tomography (PET) scans are regarded as the imaging modality of choice for the assessment of treatment response in the lymphomas because they detect treatment response when there may not be changes on anatomical imaging. Any computerized analysis of lymphomas in whole-body PET requires automatic segmentation of the studies so that sites of disease can be quantitatively monitored over time. State-of-the-art PET image segmentation methods are based on convolutional neural networks (CNNs) given their ability to leverage annotated datasets to derive high-level features about the disease process. Such methods, however, focus on PET images from a single time-point and discard information from other scans or are targeted towards specific organs and cannot cater for the multiple structures in whole-body PET images. In this study, we propose a spatio-temporal 'dual-stream' neural network (ST-DSNN) to segment sequential whole-body PET scans. Our ST-DSNN learns and accumulates image features from the PET images done over time. The accumulated image features are used to enhance the organs / structures that are consistent over time to allow easier identification of sites of active lymphoma. Our results show that our method outperforms the state-of-the-art PET image segmentation methods.
Predicting Distant Metastases in Soft-Tissue Sarcomas from PET-CT scans using Constrained Hierarchical Multi-Modality Feature Learning
Apr 23, 2021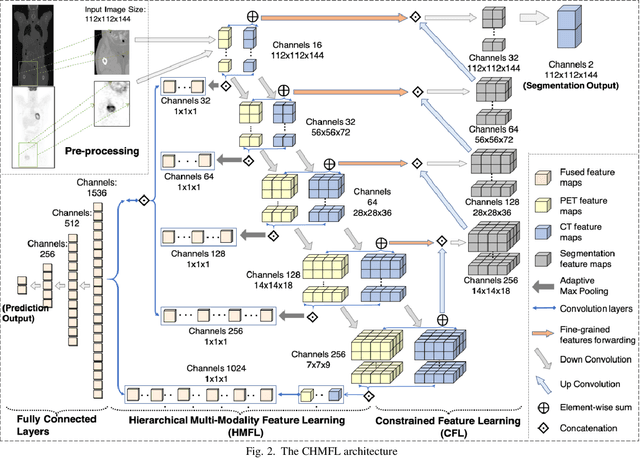
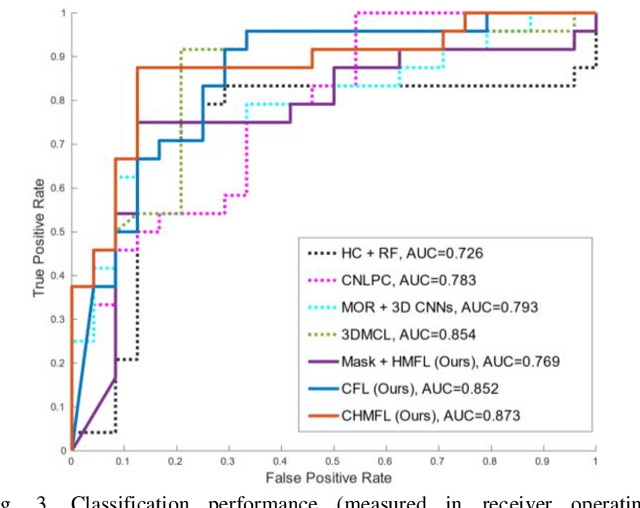
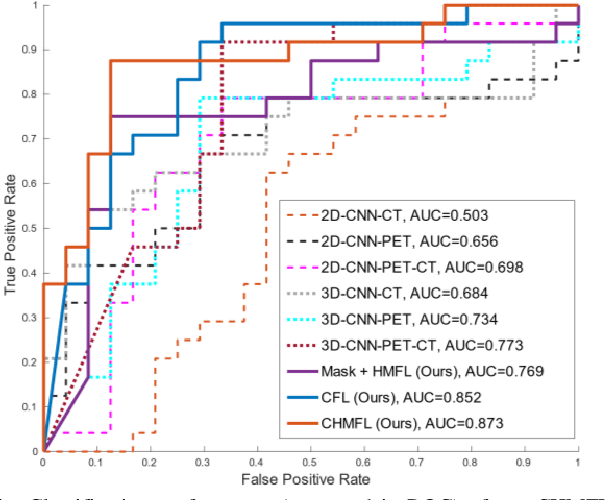
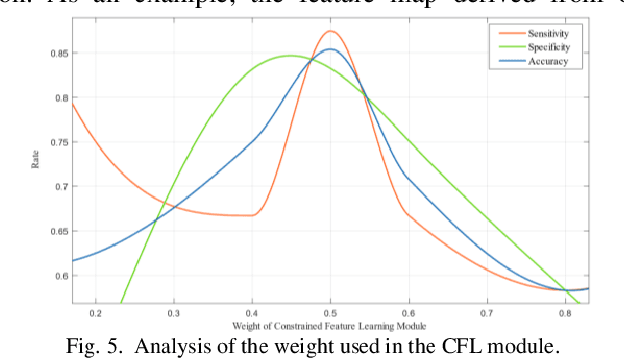
Abstract:Distant metastases (DM) refer to the dissemination of tumors, usually, beyond the organ where the tumor originated. They are the leading cause of death in patients with soft-tissue sarcomas (STSs). Positron emission tomography-computed tomography (PET-CT) is regarded as the imaging modality of choice for the management of STSs. It is difficult to determine from imaging studies which STS patients will develop metastases. 'Radiomics' refers to the extraction and analysis of quantitative features from medical images and it has been employed to help identify such tumors. The state-of-the-art in radiomics is based on convolutional neural networks (CNNs). Most CNNs are designed for single-modality imaging data (CT or PET alone) and do not exploit the information embedded in PET-CT where there is a combination of an anatomical and functional imaging modality. Furthermore, most radiomic methods rely on manual input from imaging specialists for tumor delineation, definition and selection of radiomic features. This approach, however, may not be scalable to tumors with complex boundaries and where there are multiple other sites of disease. We outline a new 3D CNN to help predict DM in STS patients from PET-CT data. The 3D CNN uses a constrained feature learning module and a hierarchical multi-modality feature learning module that leverages the complementary information from the modalities to focus on semantically important regions. Our results on a public PET-CT dataset of STS patients show that multi-modal information improves the ability to identify those patients who develop DM. Further our method outperformed all other related state-of-the-art methods.
Graph-Based Intercategory and Intermodality Network for Multilabel Classification and Melanoma Diagnosis of Skin Lesions in Dermoscopy and Clinical Images
Apr 01, 2021
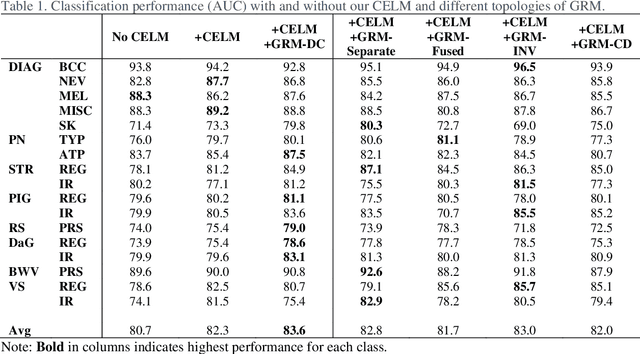
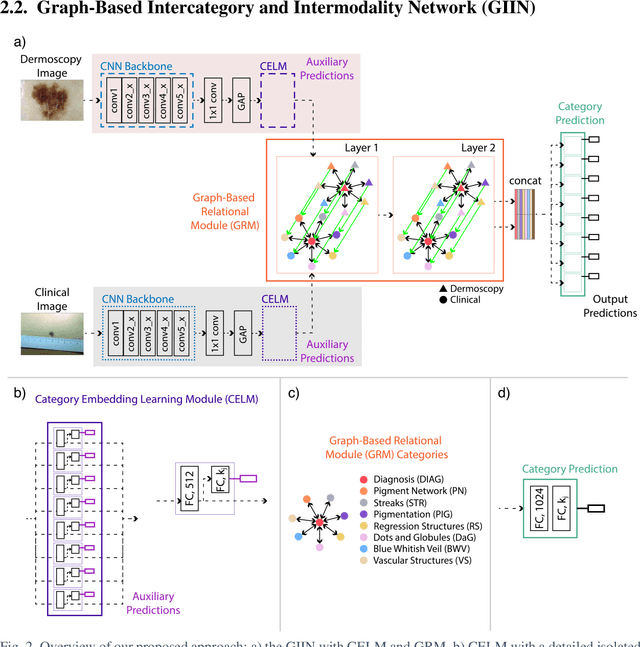
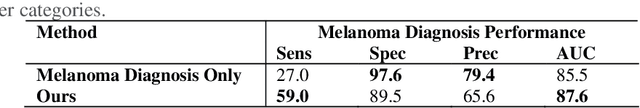
Abstract:The identification of melanoma involves an integrated analysis of skin lesion images acquired using the clinical and dermoscopy modalities. Dermoscopic images provide a detailed view of the subsurface visual structures that supplement the macroscopic clinical images. Melanoma diagnosis is commonly based on the 7-point visual category checklist (7PC). The 7PC contains intrinsic relationships between categories that can aid classification, such as shared features, correlations, and the contributions of categories towards diagnosis. Manual classification is subjective and prone to intra- and interobserver variability. This presents an opportunity for automated methods to improve diagnosis. Current state-of-the-art methods focus on a single image modality and ignore information from the other, or do not fully leverage the complementary information from both modalities. Further, there is not a method to exploit the intercategory relationships in the 7PC. In this study, we address these issues by proposing a graph-based intercategory and intermodality network (GIIN) with two modules. A graph-based relational module (GRM) leverages intercategorical relations, intermodal relations, and prioritises the visual structure details from dermoscopy by encoding category representations in a graph network. The category embedding learning module (CELM) captures representations that are specialised for each category and support the GRM. We show that our modules are effective at enhancing classification performance using a public dataset of dermoscopy-clinical images, and show that our method outperforms the state-of-the-art at classifying the 7PC categories and diagnosis.
Attention-Enhanced Cross-Task Network for Analysing Multiple Attributes of Lung Nodules in CT
Mar 05, 2021
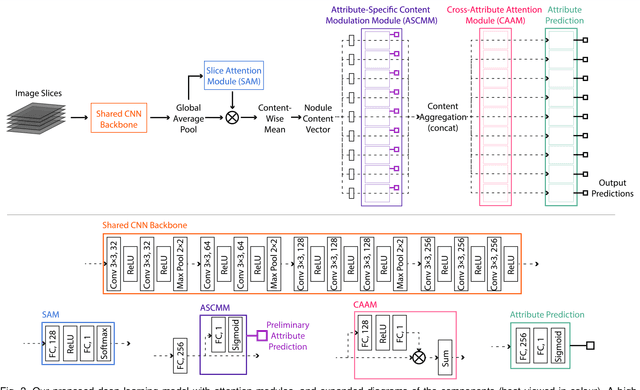
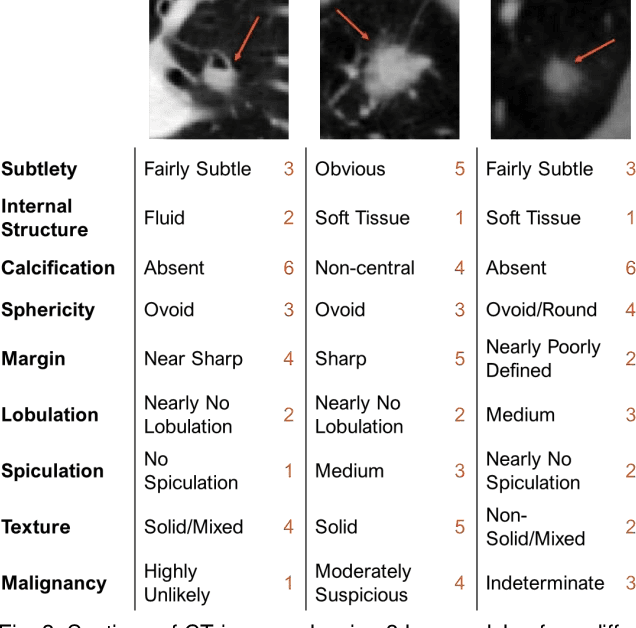
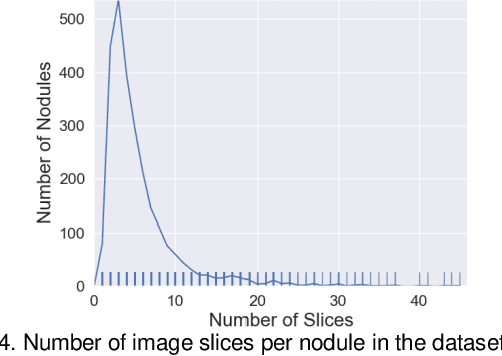
Abstract:Accurate characterisation of visual attributes such as spiculation, lobulation, and calcification of lung nodules is critical in cancer management. The characterisation of these attributes is often subjective, which may lead to high inter- and intra-observer variability. Furthermore, lung nodules are often heterogeneous in the cross-sectional image slices of a 3D volume. Current state-of-the-art methods that score multiple attributes rely on deep learning-based multi-task learning (MTL) schemes. These methods, however, extract shared visual features across attributes and then examine each attribute without explicitly leveraging their inherent intercorrelations. Furthermore, current methods either treat each slice with equal importance without considering their relevance or heterogeneity, or restrict the number of input slices, which limits performance. In this study, we address these challenges with a new convolutional neural network (CNN)-based MTL model that incorporates attention modules to simultaneously score 9 visual attributes of lung nodules in computed tomography (CT) image volumes. Our model processes entire nodule volumes of arbitrary depth and uses a slice attention module to filter out irrelevant slices. We also introduce cross-attribute and attribute specialisation attention modules that learn an optimal amalgamation of meaningful representations to leverage relationships between attributes. We demonstrate that our model outperforms previous state-of-the-art methods at scoring attributes using the well-known public LIDC-IDRI dataset of pulmonary nodules from over 1,000 patients. Our attention modules also provide easy-to-interpret weights that offer insights into the predictions of the model.
Multimodal Spatial Attention Module for Targeting Multimodal PET-CT Lung Tumor Segmentation
Aug 06, 2020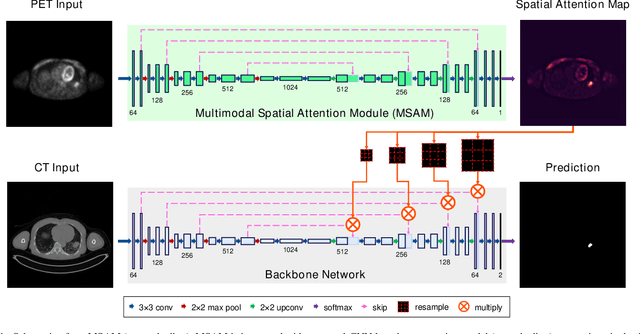
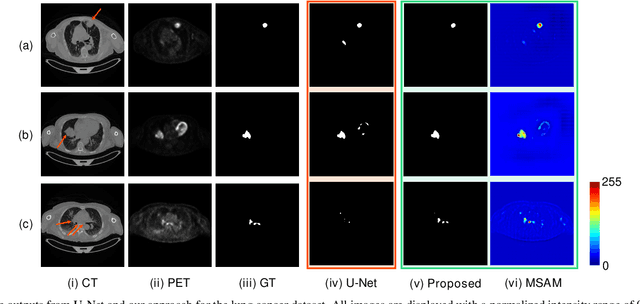
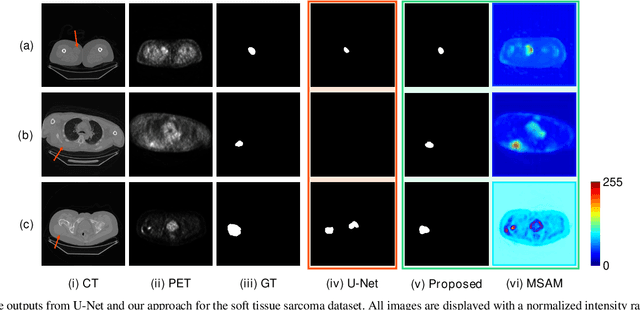
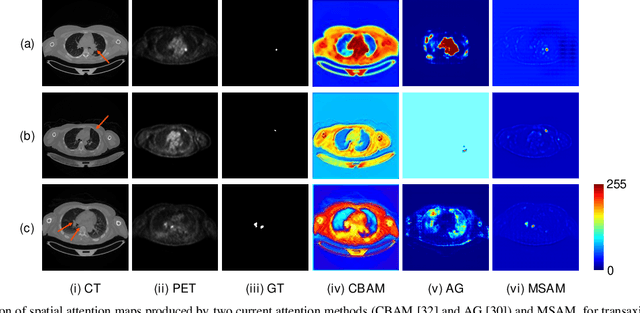
Abstract:Multimodal positron emission tomography-computed tomography (PET-CT) is used routinely in the assessment of cancer. PET-CT combines the high sensitivity for tumor detection with PET and anatomical information from CT. Tumor segmentation is a critical element of PET-CT but at present, there is not an accurate automated segmentation method. Segmentation tends to be done manually by different imaging experts and it is labor-intensive and prone to errors and inconsistency. Previous automated segmentation methods largely focused on fusing information that is extracted separately from the PET and CT modalities, with the underlying assumption that each modality contains complementary information. However, these methods do not fully exploit the high PET tumor sensitivity that can guide the segmentation. We introduce a multimodal spatial attention module (MSAM) that automatically learns to emphasize regions (spatial areas) related to tumors and suppress normal regions with physiologic high-uptake. The resulting spatial attention maps are subsequently employed to target a convolutional neural network (CNN) for segmentation of areas with higher tumor likelihood. Our MSAM can be applied to common backbone architectures and trained end-to-end. Our experimental results on two clinical PET-CT datasets of non-small cell lung cancer (NSCLC) and soft tissue sarcoma (STS) validate the effectiveness of the MSAM in these different cancer types. We show that our MSAM, with a conventional U-Net backbone, surpasses the state-of-the-art lung tumor segmentation approach by a margin of 7.6% in Dice similarity coefficient (DSC).
Semi-supervised estimation of event temporal length for cell event detection
Sep 22, 2019
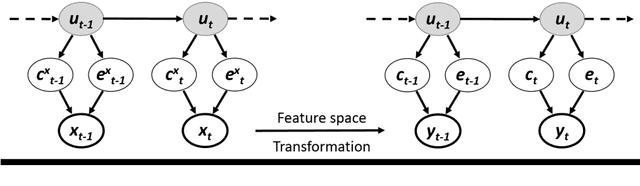
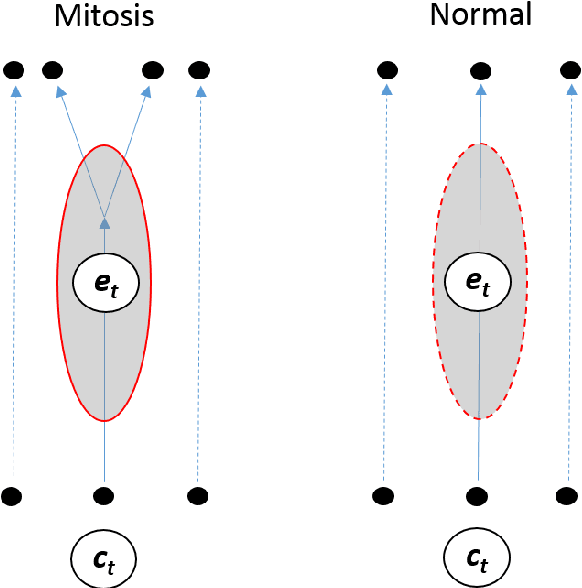
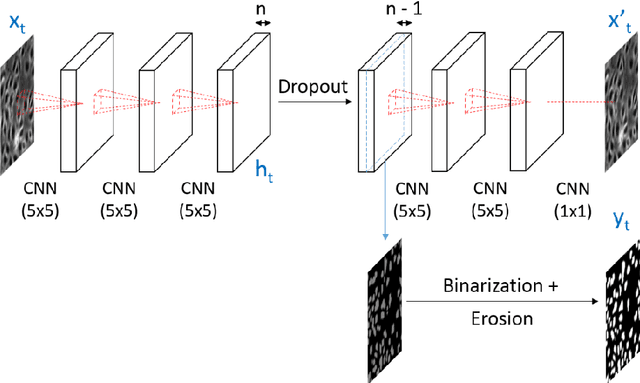
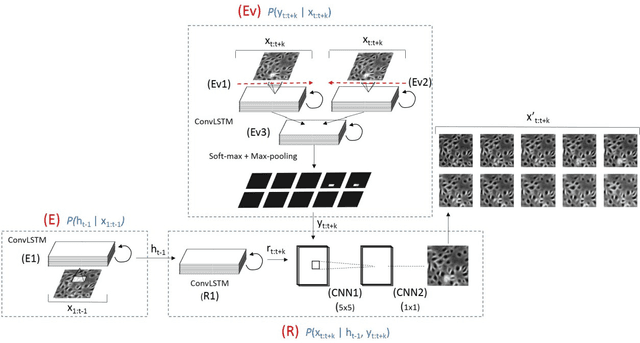
Abstract:Cell event detection in cell videos is essential for monitoring of cellular behavior over extended time periods. Deep learning methods have shown great success in the detection of cell events for their ability to capture more discriminative features of cellular processes compared to traditional methods. In particular, convolutional long short-term memory (LSTM) models, which exploits the changes in cell events observable in video sequences, is the state-of-the-art for mitosis detection in cell videos. However, their limitations are the determination of the input sequence length, which is often performed empirically, and the need for a large annotated training dataset which is expensive to prepare. We propose a novel semi-supervised method of optimal length detection for mitosis detection with two key contributions: (i) an unsupervised step for learning the spatial and temporal locations of cells in their normal stage and approximating the distribution of temporal lengths of cell events and, (ii) a step of inferring, from that distribution, an optimal input sequence length and a minimal number of annotated frames for training a LSTM model for each particular video. We evaluated our method in detecting mitosis in densely packed stem cells in a phase-contrast microscopy videos. Our experimental data prove that increasing the input sequence length of LSTM can lead to a decrease in performance. Our results also show that by approximating the optimal input sequence length of the tested video, a model trained with only 18 annotated frames achieved F1-scores of 0.880-0.907, which are 10% higher than those of other published methods with a full set of 110 training annotated frames.
Unsupervised Feature Learning with K-means and An Ensemble of Deep Convolutional Neural Networks for Medical Image Classification
Jun 07, 2019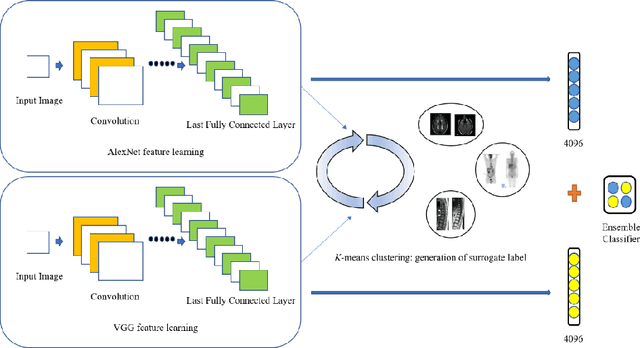
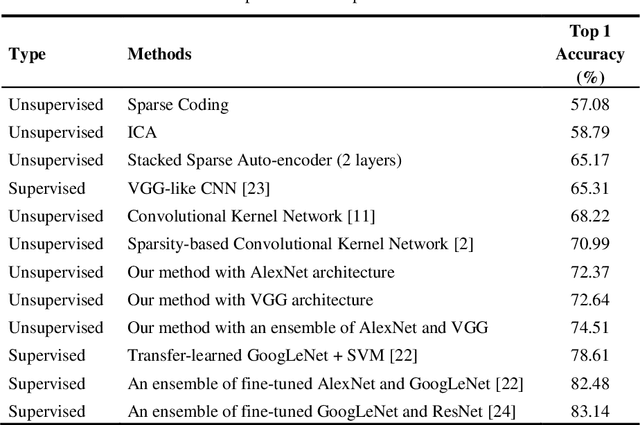
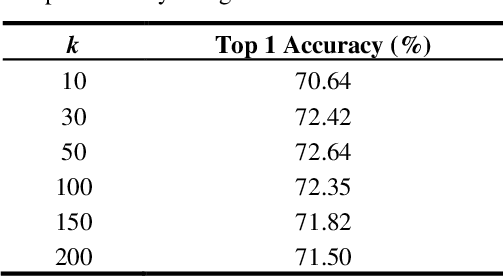
Abstract:Medical image analysis using supervised deep learning methods remains problematic because of the reliance of deep learning methods on large amounts of labelled training data. Although medical imaging data repositories continue to expand there has not been a commensurate increase in the amount of annotated data. Hence, we propose a new unsupervised feature learning method that learns feature representations to then differentiate dissimilar medical images using an ensemble of different convolutional neural networks (CNNs) and K-means clustering. It jointly learns feature representations and clustering assignments in an end-to-end fashion. We tested our approach on a public medical dataset and show its accuracy was better than state-of-the-art unsupervised feature learning methods and comparable to state-of-the-art supervised CNNs. Our findings suggest that our method could be used to tackle the issue of the large volume of unlabelled data in medical imaging repositories.
Unsupervised Deep Transfer Feature Learning for Medical Image Classification
Mar 26, 2019
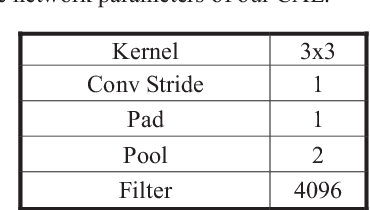
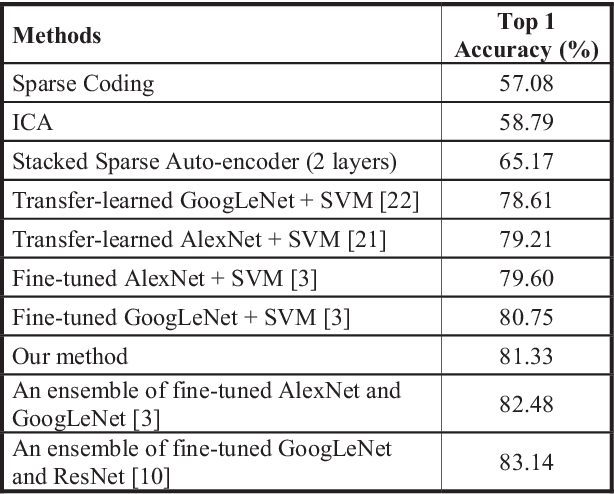
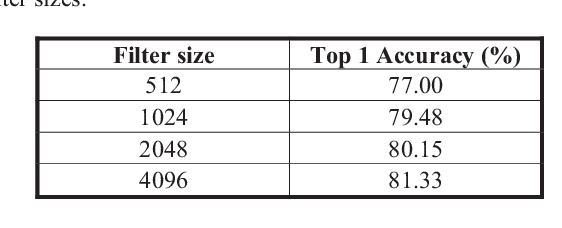
Abstract:The accuracy and robustness of image classification with supervised deep learning are dependent on the availability of large-scale, annotated training data. However, there is a paucity of annotated data available due to the complexity of manual annotation. To overcome this problem, a popular approach is to use transferable knowledge across different domains by: 1) using a generic feature extractor that has been pre-trained on large-scale general images (i.e., transfer-learned) but which not suited to capture characteristics from medical images; or 2) fine-tuning generic knowledge with a relatively smaller number of annotated images. Our aim is to reduce the reliance on annotated training data by using a new hierarchical unsupervised feature extractor with a convolutional auto-encoder placed atop of a pre-trained convolutional neural network. Our approach constrains the rich and generic image features from the pre-trained domain to a sophisticated representation of the local image characteristics from the unannotated medical image domain. Our approach has a higher classification accuracy than transfer-learned approaches and is competitive with state-of-the-art supervised fine-tuned methods.
Sparsity-based Convolutional Kernel Network for Unsupervised Medical Image Analysis
Jul 27, 2018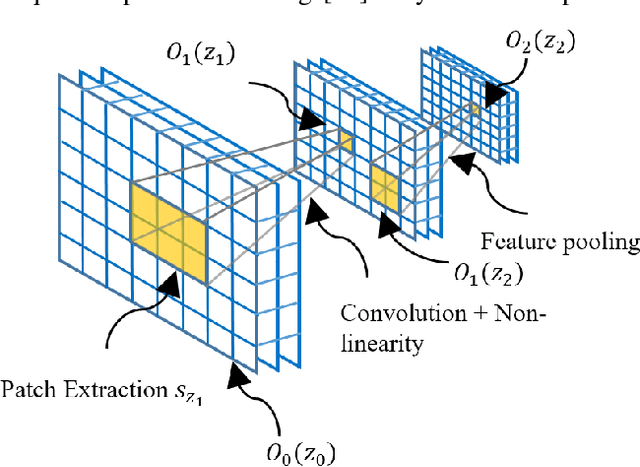
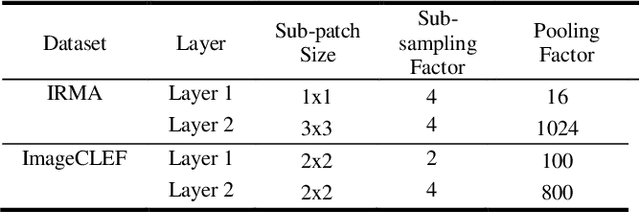
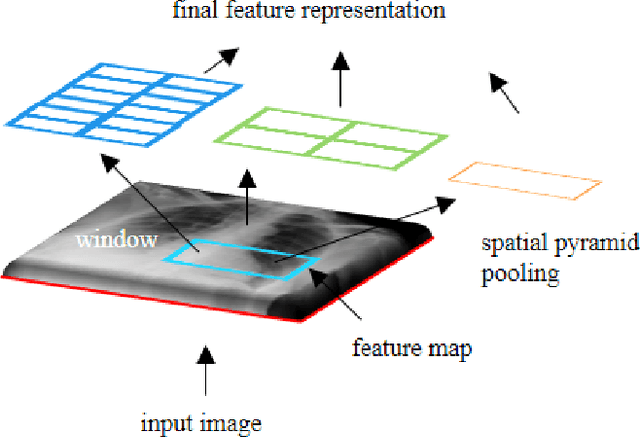
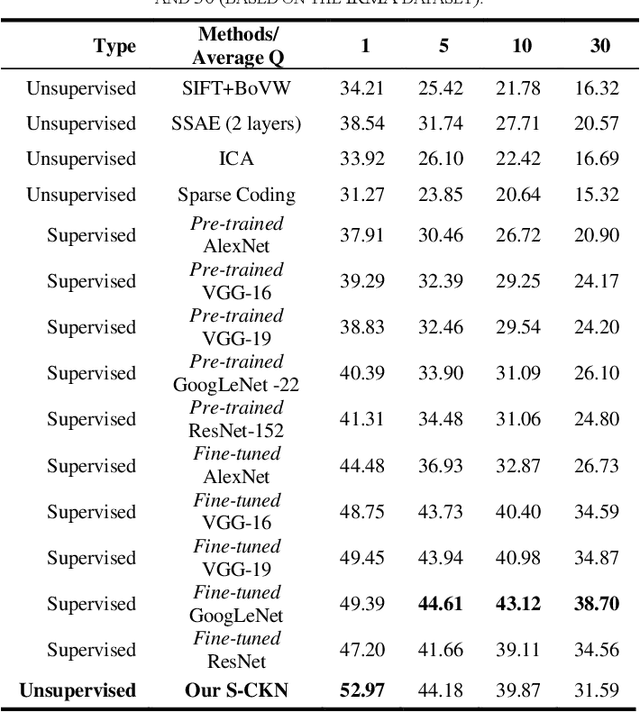
Abstract:The availability of large-scale annotated image datasets coupled with recent advances in supervised deep learning methods are enabling the derivation of representative image features that can potentially impact different image analysis problems. However, such supervised approaches are not feasible in the medical domain where it is challenging to obtain a large volume of labelled data due to the complexity of manual annotation and inter- and intra-observer variability in label assignment. Algorithms designed to work on small annotated datasets are useful but have limited applications. In an effort to address the lack of annotated data in the medical image analysis domain, we propose an algorithm for hierarchical unsupervised feature learning. Our algorithm introduces three new contributions: (i) we use kernel learning to identify and represent invariant characteristics across image sub-patches in an unsupervised manner; (ii) we leverage the sparsity inherent to medical image data and propose a new sparse convolutional kernel network (S-CKN) that can be pre-trained in a layer-wise fashion, thereby providing initial discriminative features for medical data; and (iii) we propose a spatial pyramid pooling framework to capture subtle geometric differences in medical image data. Our experiments evaluate our algorithm in two common application areas of medical image retrieval and classification using two public datasets. Our results demonstrate that the medical image feature representations extracted with our algorithm enable a higher accuracy in both application areas compared to features extracted from other conventional unsupervised methods. Furthermore, our approach achieves an accuracy that is competitive with state-of-the-art supervised CNNs.
An unsupervised long short-term memory neural network for event detection in cell videos
Sep 07, 2017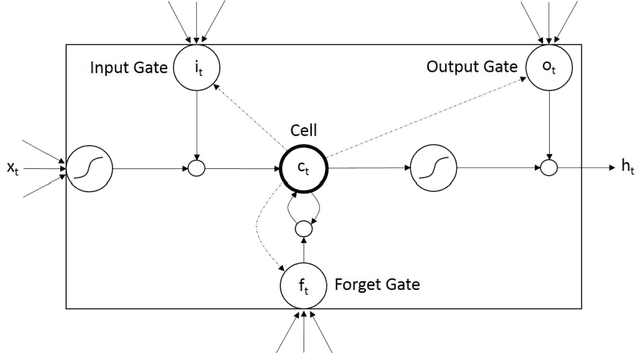
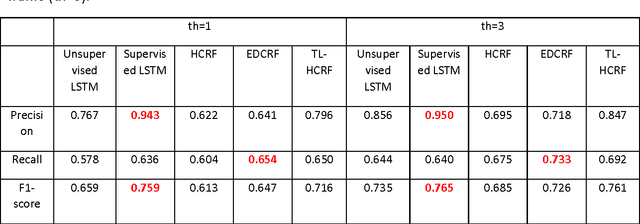
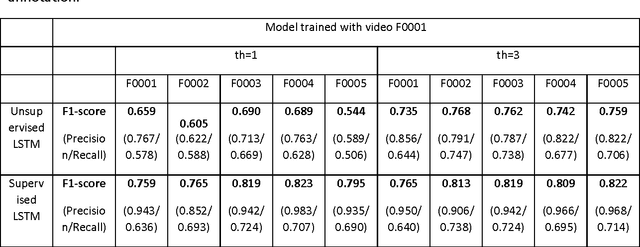
Abstract:We propose an automatic unsupervised cell event detection and classification method, which expands convolutional Long Short-Term Memory (LSTM) neural networks, for cellular events in cell video sequences. Cells in images that are captured from various biomedical applications usually have different shapes and motility, which pose difficulties for the automated event detection in cell videos. Current methods to detect cellular events are based on supervised machine learning and rely on tedious manual annotation from investigators with specific expertise. So that our LSTM network could be trained in an unsupervised manner, we designed it with a branched structure where one branch learns the frequent, regular appearance and movements of objects and the second learns the stochastic events, which occur rarely and without warning in a cell video sequence. We tested our network on a publicly available dataset of densely packed stem cell phase-contrast microscopy images undergoing cell division. This dataset is considered to be more challenging that a dataset with sparse cells. We compared our method to several published supervised methods evaluated on the same dataset and to a supervised LSTM method with a similar design and configuration to our unsupervised method. We used an F1-score, which is a balanced measure for both precision and recall. Our results show that our unsupervised method has a higher or similar F1-score when compared to two fully supervised methods that are based on Hidden Conditional Random Fields (HCRF), and has comparable accuracy with the current best supervised HCRF-based method. Our method was generalizable as after being trained on one video it could be applied to videos where the cells were in different conditions. The accuracy of our unsupervised method approached that of its supervised counterpart.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge