Sparsity-based Convolutional Kernel Network for Unsupervised Medical Image Analysis
Paper and Code
Jul 27, 2018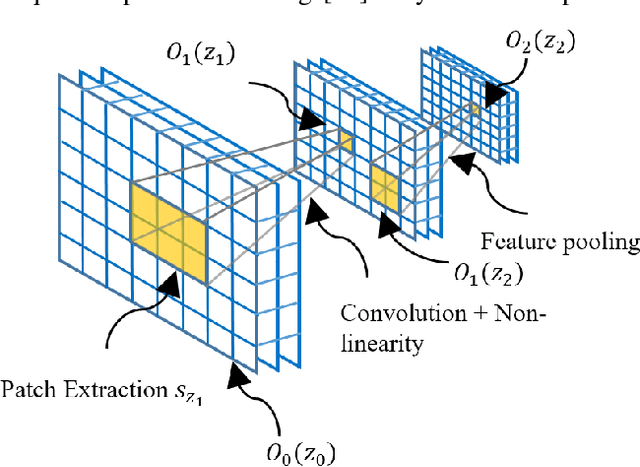
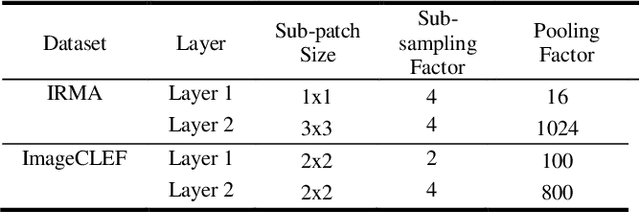
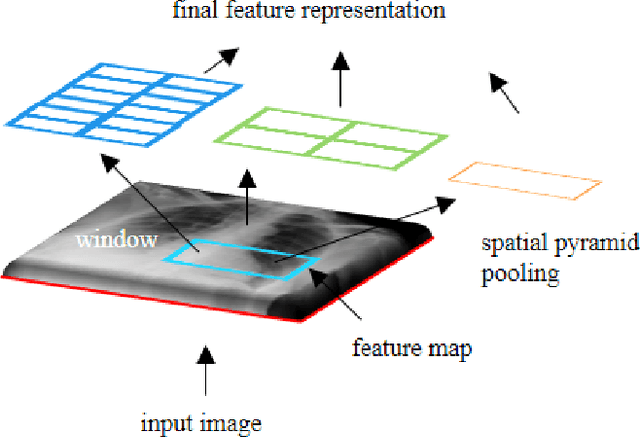
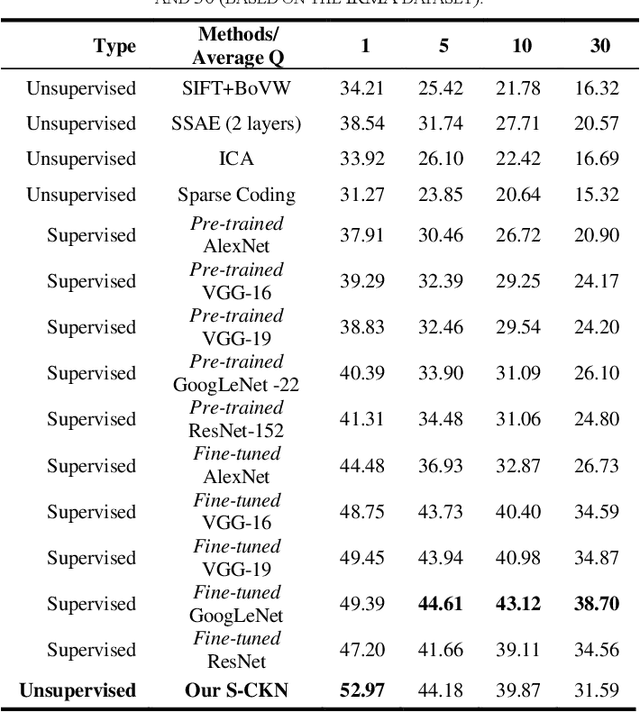
The availability of large-scale annotated image datasets coupled with recent advances in supervised deep learning methods are enabling the derivation of representative image features that can potentially impact different image analysis problems. However, such supervised approaches are not feasible in the medical domain where it is challenging to obtain a large volume of labelled data due to the complexity of manual annotation and inter- and intra-observer variability in label assignment. Algorithms designed to work on small annotated datasets are useful but have limited applications. In an effort to address the lack of annotated data in the medical image analysis domain, we propose an algorithm for hierarchical unsupervised feature learning. Our algorithm introduces three new contributions: (i) we use kernel learning to identify and represent invariant characteristics across image sub-patches in an unsupervised manner; (ii) we leverage the sparsity inherent to medical image data and propose a new sparse convolutional kernel network (S-CKN) that can be pre-trained in a layer-wise fashion, thereby providing initial discriminative features for medical data; and (iii) we propose a spatial pyramid pooling framework to capture subtle geometric differences in medical image data. Our experiments evaluate our algorithm in two common application areas of medical image retrieval and classification using two public datasets. Our results demonstrate that the medical image feature representations extracted with our algorithm enable a higher accuracy in both application areas compared to features extracted from other conventional unsupervised methods. Furthermore, our approach achieves an accuracy that is competitive with state-of-the-art supervised CNNs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge