Ruiyan Zhang
Automatic Left Ventricular Cavity Segmentation via Deep Spatial Sequential Network in 4D Computed Tomography Studies
Dec 17, 2024



Abstract:Automated segmentation of left ventricular cavity (LVC) in temporal cardiac image sequences (multiple time points) is a fundamental requirement for quantitative analysis of its structural and functional changes. Deep learning based methods for the segmentation of LVC are the state of the art; however, these methods are generally formulated to work on single time points, and fails to exploit the complementary information from the temporal image sequences that can aid in segmentation accuracy and consistency among the images across the time points. Furthermore, these segmentation methods perform poorly in segmenting the end-systole (ES) phase images, where the left ventricle deforms to the smallest irregular shape, and the boundary between the blood chamber and myocardium becomes inconspicuous. To overcome these limitations, we propose a new method to automatically segment temporal cardiac images where we introduce a spatial sequential (SS) network to learn the deformation and motion characteristics of the LVC in an unsupervised manner; these characteristics were then integrated with sequential context information derived from bi-directional learning (BL) where both chronological and reverse-chronological directions of the image sequence were used. Our experimental results on a cardiac computed tomography (CT) dataset demonstrated that our spatial-sequential network with bi-directional learning (SS-BL) method outperformed existing methods for LVC segmentation. Our method was also applied to MRI cardiac dataset and the results demonstrated the generalizability of our method.
Unsupervised Landmark Detection Based Spatiotemporal Motion Estimation for 4D Dynamic Medical Images
Oct 12, 2021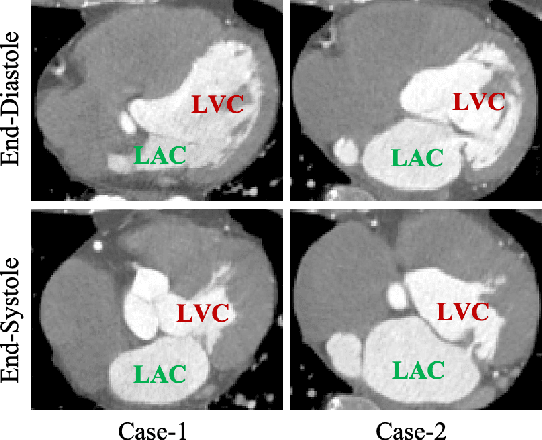
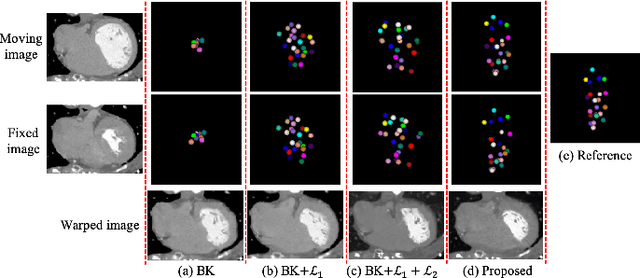
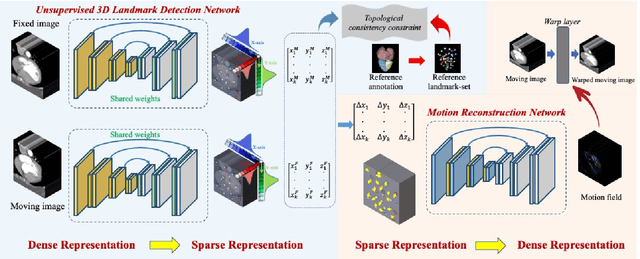
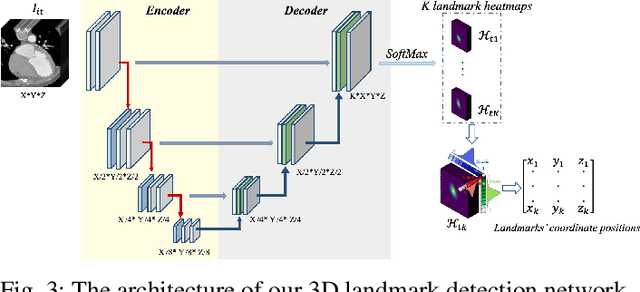
Abstract:Motion estimation is a fundamental step in dynamic medical image processing for the assessment of target organ anatomy and function. However, existing image-based motion estimation methods, which optimize the motion field by evaluating the local image similarity, are prone to produce implausible estimation, especially in the presence of large motion. In this study, we provide a novel motion estimation framework of Dense-Sparse-Dense (DSD), which comprises two stages. In the first stage, we process the raw dense image to extract sparse landmarks to represent the target organ anatomical topology and discard the redundant information that is unnecessary for motion estimation. For this purpose, we introduce an unsupervised 3D landmark detection network to extract spatially sparse but representative landmarks for the target organ motion estimation. In the second stage, we derive the sparse motion displacement from the extracted sparse landmarks of two images of different time points. Then, we present a motion reconstruction network to construct the motion field by projecting the sparse landmarks displacement back into the dense image domain. Furthermore, we employ the estimated motion field from our two-stage DSD framework as initialization and boost the motion estimation quality in light-weight yet effective iterative optimization. We evaluate our method on two dynamic medical imaging tasks to model cardiac motion and lung respiratory motion, respectively. Our method has produced superior motion estimation accuracy compared to existing comparative methods. Besides, the extensive experimental results demonstrate that our solution can extract well representative anatomical landmarks without any requirement of manual annotation. Our code is publicly available online.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge