Liyun Chen
DINO-AD: Unsupervised Anomaly Detection with Frozen DINO-V3 Features
Jan 31, 2026Abstract:Unsupervised anomaly detection (AD) in medical images aims to identify abnormal regions without relying on pixel-level annotations, which is crucial for scalable and label-efficient diagnostic systems. In this paper, we propose a novel anomaly detection framework based on DINO-V3 representations, termed DINO-AD, which leverages self-supervised visual features for precise and interpretable anomaly localization. Specifically, we introduce an embedding similarity matching strategy to select a semantically aligned support image and a foreground-aware K-means clustering module to model the distribution of normal features. Anomaly maps are then computed by comparing the query features with clustered normal embeddings through cosine similarity. Experimental results on both the Brain and Liver datasets demonstrate that our method achieves superior quantitative performance compared with state-of-the-art approaches, achieving AUROC scores of up to 98.71. Qualitative results further confirm that our framework produces clearer and more accurate anomaly localization. Extensive ablation studies validate the effectiveness of each proposed component, highlighting the robustness and generalizability of our approach.
Unsupervised Landmark Detection Based Spatiotemporal Motion Estimation for 4D Dynamic Medical Images
Oct 12, 2021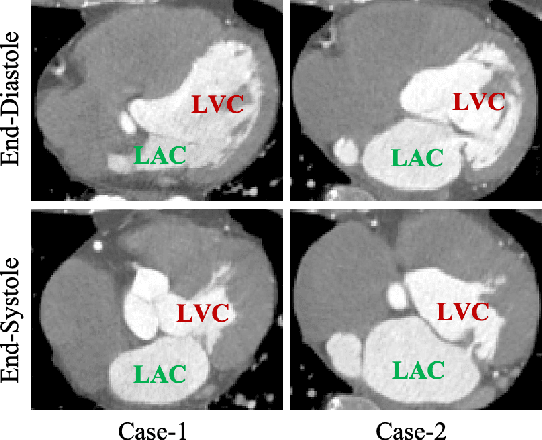
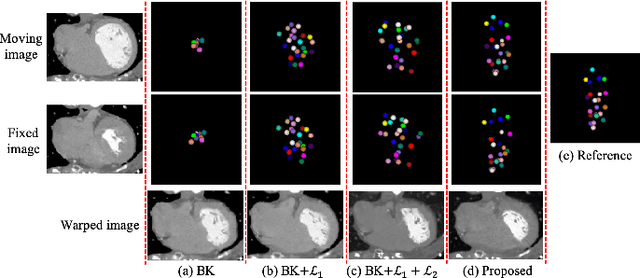
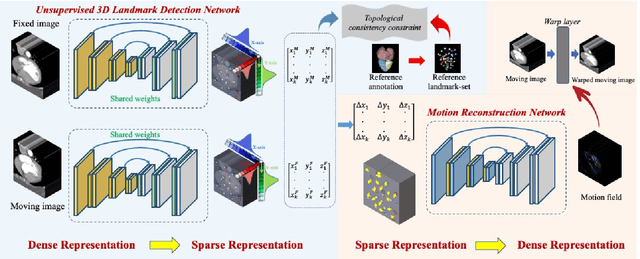
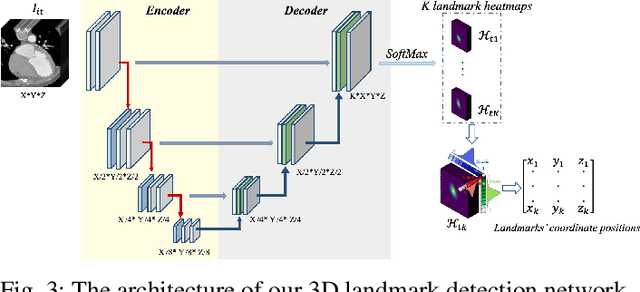
Abstract:Motion estimation is a fundamental step in dynamic medical image processing for the assessment of target organ anatomy and function. However, existing image-based motion estimation methods, which optimize the motion field by evaluating the local image similarity, are prone to produce implausible estimation, especially in the presence of large motion. In this study, we provide a novel motion estimation framework of Dense-Sparse-Dense (DSD), which comprises two stages. In the first stage, we process the raw dense image to extract sparse landmarks to represent the target organ anatomical topology and discard the redundant information that is unnecessary for motion estimation. For this purpose, we introduce an unsupervised 3D landmark detection network to extract spatially sparse but representative landmarks for the target organ motion estimation. In the second stage, we derive the sparse motion displacement from the extracted sparse landmarks of two images of different time points. Then, we present a motion reconstruction network to construct the motion field by projecting the sparse landmarks displacement back into the dense image domain. Furthermore, we employ the estimated motion field from our two-stage DSD framework as initialization and boost the motion estimation quality in light-weight yet effective iterative optimization. We evaluate our method on two dynamic medical imaging tasks to model cardiac motion and lung respiratory motion, respectively. Our method has produced superior motion estimation accuracy compared to existing comparative methods. Besides, the extensive experimental results demonstrate that our solution can extract well representative anatomical landmarks without any requirement of manual annotation. Our code is publicly available online.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge