Jiayu Huo
DINO-AD: Unsupervised Anomaly Detection with Frozen DINO-V3 Features
Jan 31, 2026Abstract:Unsupervised anomaly detection (AD) in medical images aims to identify abnormal regions without relying on pixel-level annotations, which is crucial for scalable and label-efficient diagnostic systems. In this paper, we propose a novel anomaly detection framework based on DINO-V3 representations, termed DINO-AD, which leverages self-supervised visual features for precise and interpretable anomaly localization. Specifically, we introduce an embedding similarity matching strategy to select a semantically aligned support image and a foreground-aware K-means clustering module to model the distribution of normal features. Anomaly maps are then computed by comparing the query features with clustered normal embeddings through cosine similarity. Experimental results on both the Brain and Liver datasets demonstrate that our method achieves superior quantitative performance compared with state-of-the-art approaches, achieving AUROC scores of up to 98.71. Qualitative results further confirm that our framework produces clearer and more accurate anomaly localization. Extensive ablation studies validate the effectiveness of each proposed component, highlighting the robustness and generalizability of our approach.
SAM-I2I: Unleash the Power of Segment Anything Model for Medical Image Translation
Nov 13, 2024



Abstract:Medical image translation is crucial for reducing the need for redundant and expensive multi-modal imaging in clinical field. However, current approaches based on Convolutional Neural Networks (CNNs) and Transformers often fail to capture fine-grain semantic features, resulting in suboptimal image quality. To address this challenge, we propose SAM-I2I, a novel image-to-image translation framework based on the Segment Anything Model 2 (SAM2). SAM-I2I utilizes a pre-trained image encoder to extract multiscale semantic features from the source image and a decoder, based on the mask unit attention module, to synthesize target modality images. Our experiments on multi-contrast MRI datasets demonstrate that SAM-I2I outperforms state-of-the-art methods, offering more efficient and accurate medical image translation.
Self-supervised Brain Lesion Generation for Effective Data Augmentation of Medical Images
Jun 21, 2024



Abstract:Accurate brain lesion delineation is important for planning neurosurgical treatment. Automatic brain lesion segmentation methods based on convolutional neural networks have demonstrated remarkable performance. However, neural network performance is constrained by the lack of large-scale well-annotated training datasets. In this manuscript, we propose a comprehensive framework to efficiently generate new, realistic samples for training a brain lesion segmentation model. We first train a lesion generator, based on an adversarial autoencoder, in a self-supervised manner. Next, we utilize a novel image composition algorithm, Soft Poisson Blending, to seamlessly combine synthetic lesions and brain images to obtain training samples. Finally, to effectively train the brain lesion segmentation model with augmented images we introduce a new prototype consistence regularization to align real and synthetic features. Our framework is validated by extensive experiments on two public brain lesion segmentation datasets: ATLAS v2.0 and Shift MS. Our method outperforms existing brain image data augmentation schemes. For instance, our method improves the Dice from 50.36% to 60.23% compared to the U-Net with conventional data augmentation techniques for the ATLAS v2.0 dataset.
A Robust Ensemble Algorithm for Ischemic Stroke Lesion Segmentation: Generalizability and Clinical Utility Beyond the ISLES Challenge
Apr 03, 2024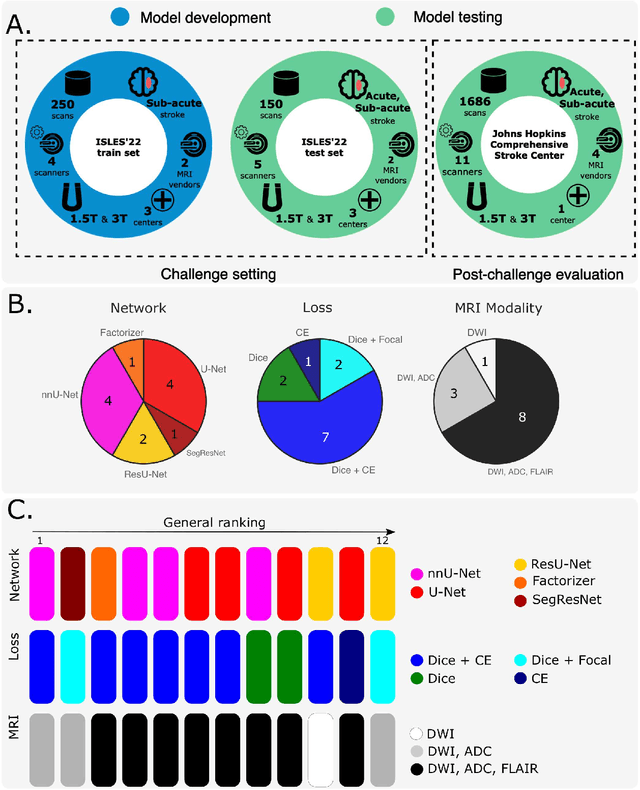
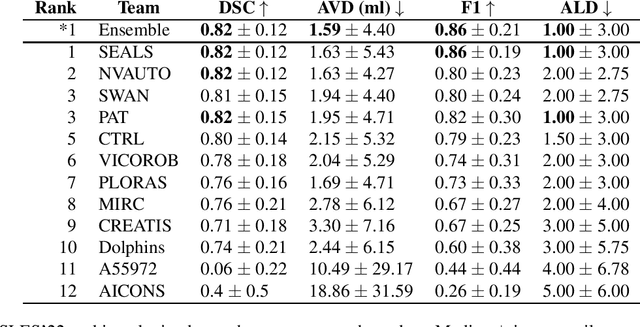
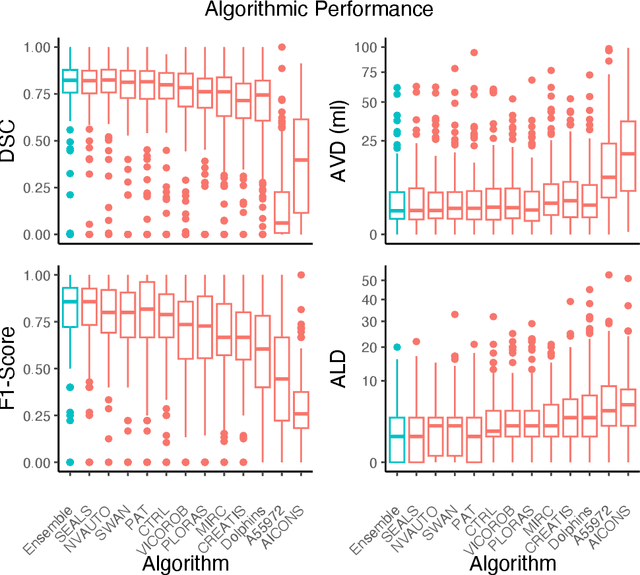
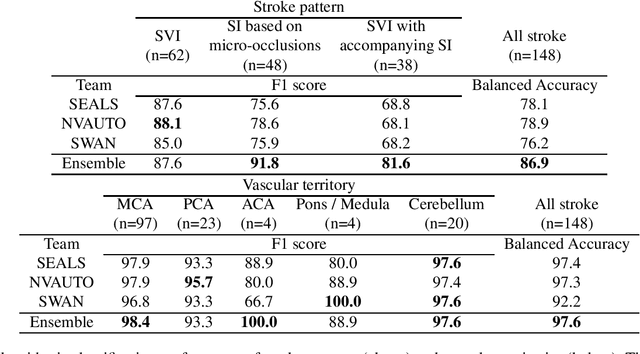
Abstract:Diffusion-weighted MRI (DWI) is essential for stroke diagnosis, treatment decisions, and prognosis. However, image and disease variability hinder the development of generalizable AI algorithms with clinical value. We address this gap by presenting a novel ensemble algorithm derived from the 2022 Ischemic Stroke Lesion Segmentation (ISLES) challenge. ISLES'22 provided 400 patient scans with ischemic stroke from various medical centers, facilitating the development of a wide range of cutting-edge segmentation algorithms by the research community. Through collaboration with leading teams, we combined top-performing algorithms into an ensemble model that overcomes the limitations of individual solutions. Our ensemble model achieved superior ischemic lesion detection and segmentation accuracy on our internal test set compared to individual algorithms. This accuracy generalized well across diverse image and disease variables. Furthermore, the model excelled in extracting clinical biomarkers. Notably, in a Turing-like test, neuroradiologists consistently preferred the algorithm's segmentations over manual expert efforts, highlighting increased comprehensiveness and precision. Validation using a real-world external dataset (N=1686) confirmed the model's generalizability. The algorithm's outputs also demonstrated strong correlations with clinical scores (admission NIHSS and 90-day mRS) on par with or exceeding expert-derived results, underlining its clinical relevance. This study offers two key findings. First, we present an ensemble algorithm (https://github.com/Tabrisrei/ISLES22_Ensemble) that detects and segments ischemic stroke lesions on DWI across diverse scenarios on par with expert (neuro)radiologists. Second, we show the potential for biomedical challenge outputs to extend beyond the challenge's initial objectives, demonstrating their real-world clinical applicability.
Generative Medical Segmentation
Mar 27, 2024



Abstract:Rapid advancements in medical image segmentation performance have been significantly driven by the development of Convolutional Neural Networks (CNNs) and Vision Transformers (ViTs). However, these models introduce high computational demands and often have limited ability to generalize across diverse medical imaging datasets. In this manuscript, we introduce Generative Medical Segmentation (GMS), a novel approach leveraging a generative model for image segmentation. Concretely, GMS employs a robust pre-trained Variational Autoencoder (VAE) to derive latent representations of both images and masks, followed by a mapping model that learns the transition from image to mask in the latent space. This process culminates in generating a precise segmentation mask within the image space using the pre-trained VAE decoder. The design of GMS leads to fewer learnable parameters in the model, resulting in a reduced computational burden and enhanced generalization capability. Our extensive experimental analysis across five public datasets in different medical imaging domains demonstrates GMS outperforms existing discriminative segmentation models and has remarkable domain generalization. Our experiments suggest GMS could set a new benchmark for medical image segmentation, offering a scalable and effective solution. GMS implementation and model weights are available at https://github.com/King-HAW/GMS.
MatchSeg: Towards Better Segmentation via Reference Image Matching
Mar 23, 2024



Abstract:Recently, automated medical image segmentation methods based on deep learning have achieved great success. However, they heavily rely on large annotated datasets, which are costly and time-consuming to acquire. Few-shot learning aims to overcome the need for annotated data by using a small labeled dataset, known as a support set, to guide predicting labels for new, unlabeled images, known as the query set. Inspired by this paradigm, we introduce MatchSeg, a novel framework that enhances medical image segmentation through strategic reference image matching. We leverage contrastive language-image pre-training (CLIP) to select highly relevant samples when defining the support set. Additionally, we design a joint attention module to strengthen the interaction between support and query features, facilitating a more effective knowledge transfer between support and query sets. We validated our method across four public datasets. Experimental results demonstrate superior segmentation performance and powerful domain generalization ability of MatchSeg against existing methods for domain-specific and cross-domain segmentation tasks. Our code is made available at https://github.com/keeplearning-again/MatchSeg
RetiGen: A Framework for Generalized Retinal Diagnosis Using Multi-View Fundus Images
Mar 22, 2024



Abstract:This study introduces a novel framework for enhancing domain generalization in medical imaging, specifically focusing on utilizing unlabelled multi-view colour fundus photographs. Unlike traditional approaches that rely on single-view imaging data and face challenges in generalizing across diverse clinical settings, our method leverages the rich information in the unlabelled multi-view imaging data to improve model robustness and accuracy. By incorporating a class balancing method, a test-time adaptation technique and a multi-view optimization strategy, we address the critical issue of domain shift that often hampers the performance of machine learning models in real-world applications. Experiments comparing various state-of-the-art domain generalization and test-time optimization methodologies show that our approach consistently outperforms when combined with existing baseline and state-of-the-art methods. We also show our online method improves all existing techniques. Our framework demonstrates improvements in domain generalization capabilities and offers a practical solution for real-world deployment by facilitating online adaptation to new, unseen datasets. Our code is available at https://github.com/zgy600/RetiGen .
DDSB: An Unsupervised and Training-free Method for Phase Detection in Echocardiography
Mar 19, 2024



Abstract:Accurate identification of End-Diastolic (ED) and End-Systolic (ES) frames is key for cardiac function assessment through echocardiography. However, traditional methods face several limitations: they require extensive amounts of data, extensive annotations by medical experts, significant training resources, and often lack robustness. Addressing these challenges, we proposed an unsupervised and training-free method, our novel approach leverages unsupervised segmentation to enhance fault tolerance against segmentation inaccuracies. By identifying anchor points and analyzing directional deformation, we effectively reduce dependence on the accuracy of initial segmentation images and enhance fault tolerance, all while improving robustness. Tested on Echo-dynamic and CAMUS datasets, our method achieves comparable accuracy to learning-based models without their associated drawbacks. The code is available at https://github.com/MRUIL/DDSB
Rethinking Low-quality Optical Flow in Unsupervised Surgical Instrument Segmentation
Mar 15, 2024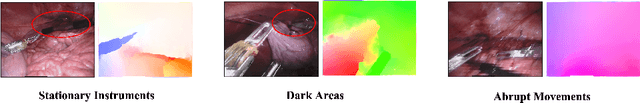
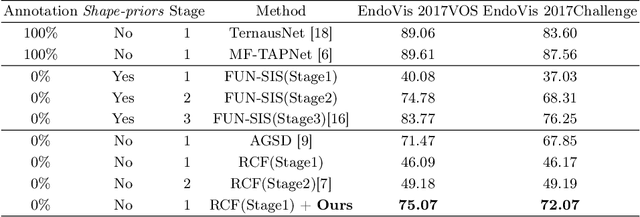
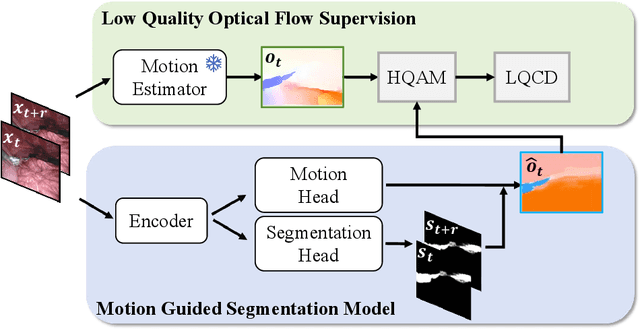
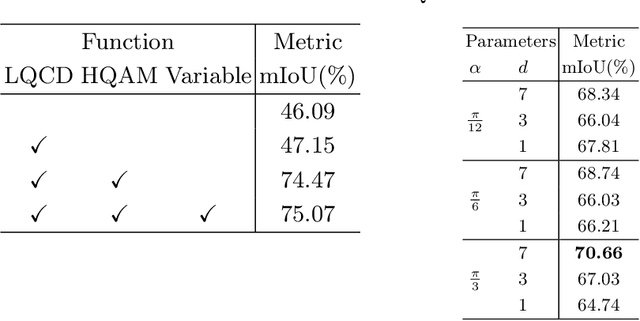
Abstract:Video-based surgical instrument segmentation plays an important role in robot-assisted surgeries. Unlike supervised settings, unsupervised segmentation relies heavily on motion cues, which are challenging to discern due to the typically lower quality of optical flow in surgical footage compared to natural scenes. This presents a considerable burden for the advancement of unsupervised segmentation techniques. In our work, we address the challenge of enhancing model performance despite the inherent limitations of low-quality optical flow. Our methodology employs a three-pronged approach: extracting boundaries directly from the optical flow, selectively discarding frames with inferior flow quality, and employing a fine-tuning process with variable frame rates. We thoroughly evaluate our strategy on the EndoVis2017 VOS dataset and Endovis2017 Challenge dataset, where our model demonstrates promising results, achieving a mean Intersection-over-Union (mIoU) of 0.75 and 0.72, respectively. Our findings suggest that our approach can greatly decrease the need for manual annotations in clinical environments and may facilitate the annotation process for new datasets. The code is available at https://github.com/wpr1018001/Rethinking-Low-quality-Optical-Flow.git
SAR-RARP50: Segmentation of surgical instrumentation and Action Recognition on Robot-Assisted Radical Prostatectomy Challenge
Dec 31, 2023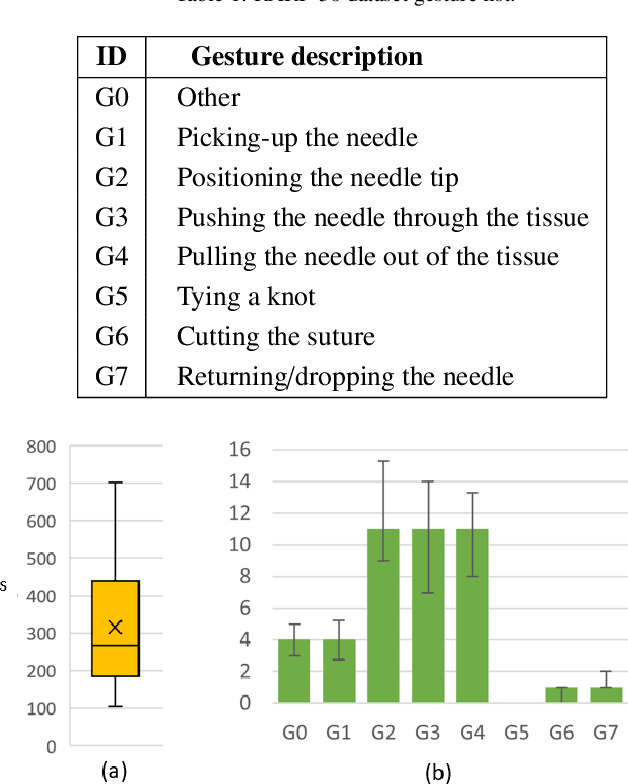
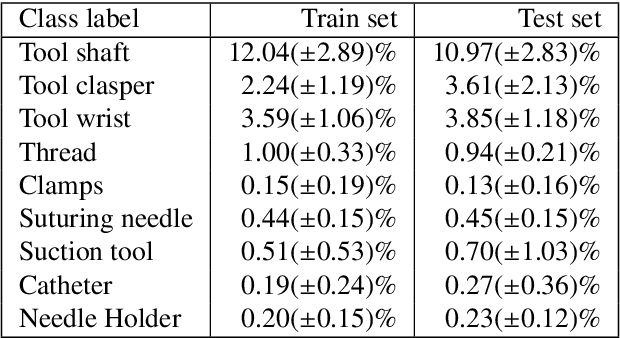
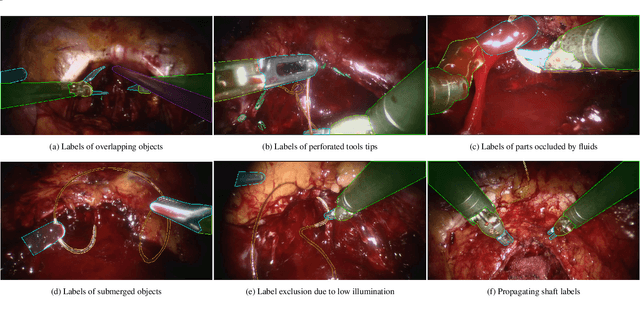
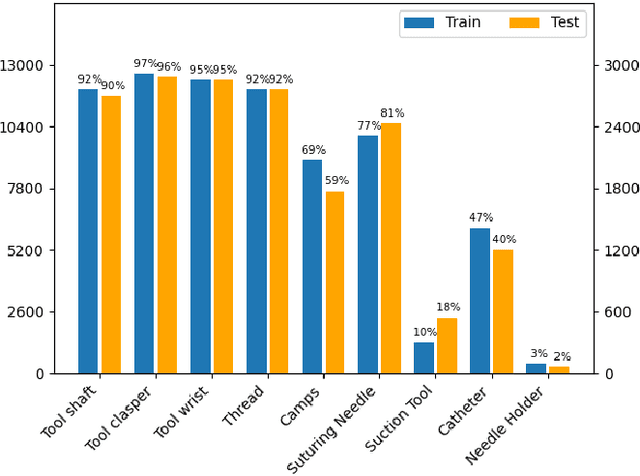
Abstract:Surgical tool segmentation and action recognition are fundamental building blocks in many computer-assisted intervention applications, ranging from surgical skills assessment to decision support systems. Nowadays, learning-based action recognition and segmentation approaches outperform classical methods, relying, however, on large, annotated datasets. Furthermore, action recognition and tool segmentation algorithms are often trained and make predictions in isolation from each other, without exploiting potential cross-task relationships. With the EndoVis 2022 SAR-RARP50 challenge, we release the first multimodal, publicly available, in-vivo, dataset for surgical action recognition and semantic instrumentation segmentation, containing 50 suturing video segments of Robotic Assisted Radical Prostatectomy (RARP). The aim of the challenge is twofold. First, to enable researchers to leverage the scale of the provided dataset and develop robust and highly accurate single-task action recognition and tool segmentation approaches in the surgical domain. Second, to further explore the potential of multitask-based learning approaches and determine their comparative advantage against their single-task counterparts. A total of 12 teams participated in the challenge, contributing 7 action recognition methods, 9 instrument segmentation techniques, and 4 multitask approaches that integrated both action recognition and instrument segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge