Kaini Wang
EndoAgent: A Memory-Guided Reflective Agent for Intelligent Endoscopic Vision-to-Decision Reasoning
Aug 10, 2025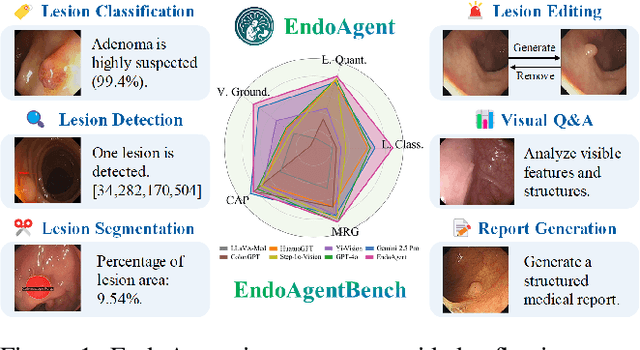
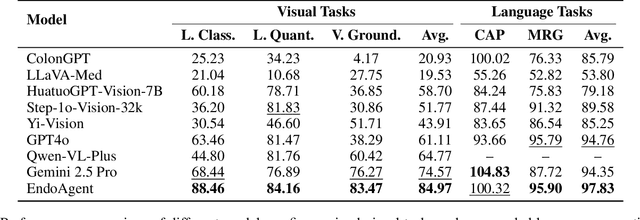
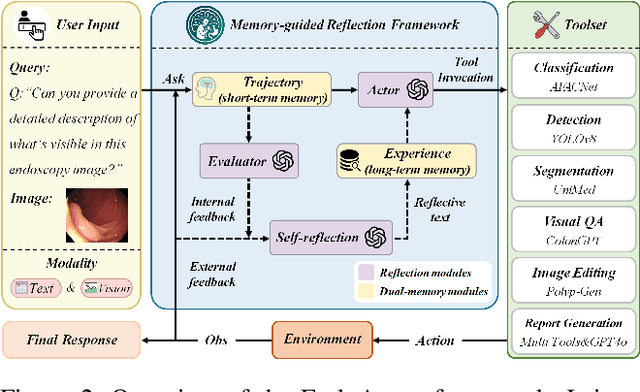
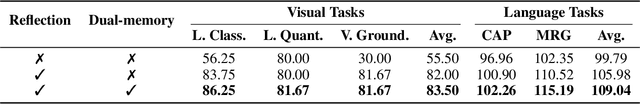
Abstract:Developing general artificial intelligence (AI) systems to support endoscopic image diagnosis is an emerging research priority. Existing methods based on large-scale pretraining often lack unified coordination across tasks and struggle to handle the multi-step processes required in complex clinical workflows. While AI agents have shown promise in flexible instruction parsing and tool integration across domains, their potential in endoscopy remains underexplored. To address this gap, we propose EndoAgent, the first memory-guided agent for vision-to-decision endoscopic analysis that integrates iterative reasoning with adaptive tool selection and collaboration. Built on a dual-memory design, it enables sophisticated decision-making by ensuring logical coherence through short-term action tracking and progressively enhancing reasoning acuity through long-term experiential learning. To support diverse clinical tasks, EndoAgent integrates a suite of expert-designed tools within a unified reasoning loop. We further introduce EndoAgentBench, a benchmark of 5,709 visual question-answer pairs that assess visual understanding and language generation capabilities in realistic scenarios. Extensive experiments show that EndoAgent consistently outperforms both general and medical multimodal models, exhibiting its strong flexibility and reasoning capabilities.
Efficient MedSAMs: Segment Anything in Medical Images on Laptop
Dec 20, 2024


Abstract:Promptable segmentation foundation models have emerged as a transformative approach to addressing the diverse needs in medical images, but most existing models require expensive computing, posing a big barrier to their adoption in clinical practice. In this work, we organized the first international competition dedicated to promptable medical image segmentation, featuring a large-scale dataset spanning nine common imaging modalities from over 20 different institutions. The top teams developed lightweight segmentation foundation models and implemented an efficient inference pipeline that substantially reduced computational requirements while maintaining state-of-the-art segmentation accuracy. Moreover, the post-challenge phase advanced the algorithms through the design of performance booster and reproducibility tasks, resulting in improved algorithms and validated reproducibility of the winning solution. Furthermore, the best-performing algorithms have been incorporated into the open-source software with a user-friendly interface to facilitate clinical adoption. The data and code are publicly available to foster the further development of medical image segmentation foundation models and pave the way for impactful real-world applications.
TSdetector: Temporal-Spatial Self-correction Collaborative Learning for Colonoscopy Video Detection
Sep 30, 2024



Abstract:CNN-based object detection models that strike a balance between performance and speed have been gradually used in polyp detection tasks. Nevertheless, accurately locating polyps within complex colonoscopy video scenes remains challenging since existing methods ignore two key issues: intra-sequence distribution heterogeneity and precision-confidence discrepancy. To address these challenges, we propose a novel Temporal-Spatial self-correction detector (TSdetector), which first integrates temporal-level consistency learning and spatial-level reliability learning to detect objects continuously. Technically, we first propose a global temporal-aware convolution, assembling the preceding information to dynamically guide the current convolution kernel to focus on global features between sequences. In addition, we designed a hierarchical queue integration mechanism to combine multi-temporal features through a progressive accumulation manner, fully leveraging contextual consistency information together with retaining long-sequence-dependency features. Meanwhile, at the spatial level, we advance a position-aware clustering to explore the spatial relationships among candidate boxes for recalibrating prediction confidence adaptively, thus eliminating redundant bounding boxes efficiently. The experimental results on three publicly available polyp video dataset show that TSdetector achieves the highest polyp detection rate and outperforms other state-of-the-art methods. The code can be available at https://github.com/soleilssss/TSdetector.
Universal Medical Image Representation Learning with Compositional Decoders
Sep 30, 2024



Abstract:Visual-language models have advanced the development of universal models, yet their application in medical imaging remains constrained by specific functional requirements and the limited data. Current general-purpose models are typically designed with task-specific branches and heads, which restricts the shared feature space and the flexibility of model. To address these challenges, we have developed a decomposed-composed universal medical imaging paradigm (UniMed) that supports tasks at all levels. To this end, we first propose a decomposed decoder that can predict two types of outputs -- pixel and semantic, based on a defined input queue. Additionally, we introduce a composed decoder that unifies the input and output spaces and standardizes task annotations across different levels into a discrete token format. The coupled design of these two components enables the model to flexibly combine tasks and mutual benefits. Moreover, our joint representation learning strategy skilfully leverages large amounts of unlabeled data and unsupervised loss, achieving efficient one-stage pretraining for more robust performance. Experimental results show that UniMed achieves state-of-the-art performance on eight datasets across all three tasks and exhibits strong zero-shot and 100-shot transferability. We will release the code and trained models upon the paper's acceptance.
DDSB: An Unsupervised and Training-free Method for Phase Detection in Echocardiography
Mar 19, 2024



Abstract:Accurate identification of End-Diastolic (ED) and End-Systolic (ES) frames is key for cardiac function assessment through echocardiography. However, traditional methods face several limitations: they require extensive amounts of data, extensive annotations by medical experts, significant training resources, and often lack robustness. Addressing these challenges, we proposed an unsupervised and training-free method, our novel approach leverages unsupervised segmentation to enhance fault tolerance against segmentation inaccuracies. By identifying anchor points and analyzing directional deformation, we effectively reduce dependence on the accuracy of initial segmentation images and enhance fault tolerance, all while improving robustness. Tested on Echo-dynamic and CAMUS datasets, our method achieves comparable accuracy to learning-based models without their associated drawbacks. The code is available at https://github.com/MRUIL/DDSB
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge