Lichi Zhang
LGAN: An Efficient High-Order Graph Neural Network via the Line Graph Aggregation
Dec 11, 2025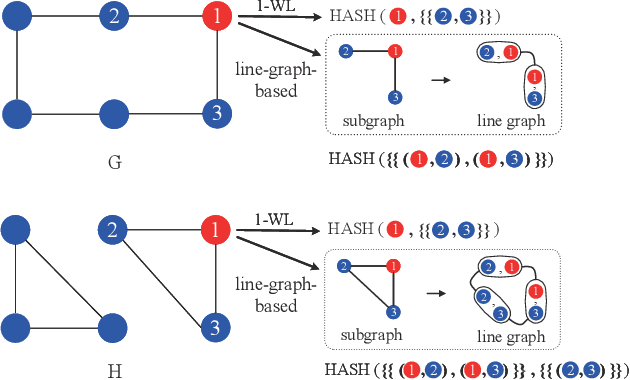
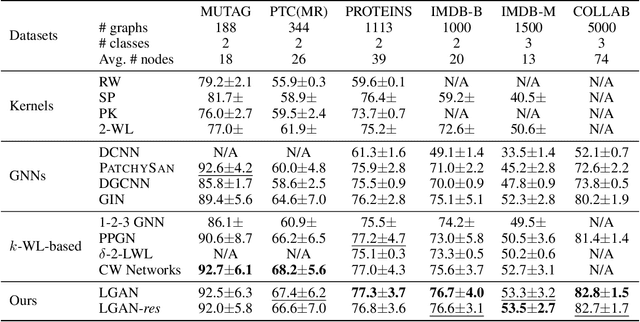
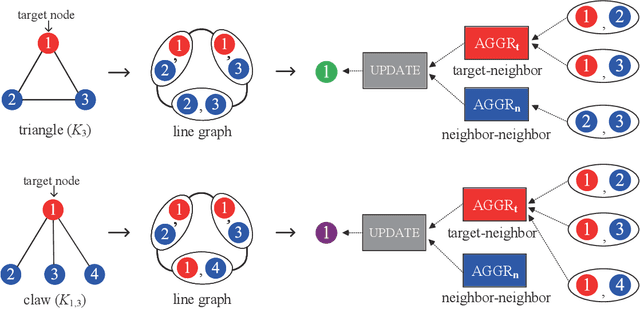
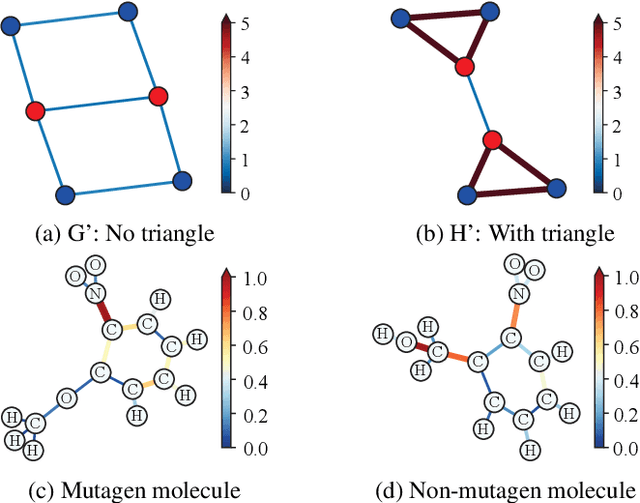
Abstract:Graph Neural Networks (GNNs) have emerged as a dominant paradigm for graph classification. Specifically, most existing GNNs mainly rely on the message passing strategy between neighbor nodes, where the expressivity is limited by the 1-dimensional Weisfeiler-Lehman (1-WL) test. Although a number of k-WL-based GNNs have been proposed to overcome this limitation, their computational cost increases rapidly with k, significantly restricting the practical applicability. Moreover, since the k-WL models mainly operate on node tuples, these k-WL-based GNNs cannot retain fine-grained node- or edge-level semantics required by attribution methods (e.g., Integrated Gradients), leading to the less interpretable problem. To overcome the above shortcomings, in this paper, we propose a novel Line Graph Aggregation Network (LGAN), that constructs a line graph from the induced subgraph centered at each node to perform the higher-order aggregation. We theoretically prove that the LGAN not only possesses the greater expressive power than the 2-WL under injective aggregation assumptions, but also has lower time complexity. Empirical evaluations on benchmarks demonstrate that the LGAN outperforms state-of-the-art k-WL-based GNNs, while offering better interpretability.
Pathology Image Restoration via Mixture of Prompts
Mar 16, 2025Abstract:In digital pathology, acquiring all-in-focus images is essential to high-quality imaging and high-efficient clinical workflow. Traditional scanners achieve this by scanning at multiple focal planes of varying depths and then merging them, which is relatively slow and often struggles with complex tissue defocus. Recent prevailing image restoration technique provides a means to restore high-quality pathology images from scans of single focal planes. However, existing image restoration methods are inadequate, due to intricate defocus patterns in pathology images and their domain-specific semantic complexities. In this work, we devise a two-stage restoration solution cascading a transformer and a diffusion model, to benefit from their powers in preserving image fidelity and perceptual quality, respectively. We particularly propose a novel mixture of prompts for the two-stage solution. Given initial prompt that models defocus in microscopic imaging, we design two prompts that describe the high-level image semantics from pathology foundation model and the fine-grained tissue structures via edge extraction. We demonstrate that, by feeding the prompt mixture to our method, we can restore high-quality pathology images from single-focal-plane scans, implying high potentials of the mixture of prompts to clinical usage. Code will be publicly available at https://github.com/caijd2000/MoP.
LSA: Latent Style Augmentation Towards Stain-Agnostic Cervical Cancer Screening
Mar 09, 2025Abstract:The deployment of computer-aided diagnosis systems for cervical cancer screening using whole slide images (WSIs) faces critical challenges due to domain shifts caused by staining variations across different scanners and imaging environments. While existing stain augmentation methods improve patch-level robustness, they fail to scale to WSIs due to two key limitations: (1) inconsistent stain patterns when extending patch operations to gigapixel slides, and (2) prohibitive computational/storage costs from offline processing of augmented WSIs.To address this, we propose Latent Style Augmentation (LSA), a framework that performs efficient, online stain augmentation directly on WSI-level latent features. We first introduce WSAug, a WSI-level stain augmentation method ensuring consistent stain across patches within a WSI. Using offline-augmented WSIs by WSAug, we design and train Stain Transformer, which can simulate targeted style in the latent space, efficiently enhancing the robustness of the WSI-level classifier. We validate our method on a multi-scanner WSI dataset for cervical cancer diagnosis. Despite being trained on data from a single scanner, our approach achieves significant performance improvements on out-of-distribution data from other scanners. Code will be available at https://github.com/caijd2000/LSA.
Structure-Guided MR-to-CT Synthesis with Spatial and Semantic Alignments for Attenuation Correction of Whole-Body PET/MR Imaging
Nov 26, 2024Abstract:Deep-learning-based MR-to-CT synthesis can estimate the electron density of tissues, thereby facilitating PET attenuation correction in whole-body PET/MR imaging. However, whole-body MR-to-CT synthesis faces several challenges including the issue of spatial misalignment and the complexity of intensity mapping, primarily due to the variety of tissues and organs throughout the whole body. Here we propose a novel whole-body MR-to-CT synthesis framework, which consists of three novel modules to tackle these challenges: (1) Structure-Guided Synthesis module leverages structure-guided attention gates to enhance synthetic image quality by diminishing unnecessary contours of soft tissues; (2) Spatial Alignment module yields precise registration between paired MR and CT images by taking into account the impacts of tissue volumes and respiratory movements, thus providing well-aligned ground-truth CT images during training; (3) Semantic Alignment module utilizes contrastive learning to constrain organ-related semantic information, thereby ensuring the semantic authenticity of synthetic CT images.We conduct extensive experiments to demonstrate that the proposed whole-body MR-to-CT framework can produce visually plausible and semantically realistic CT images, and validate its utility in PET attenuation correction.
REHRSeg: Unleashing the Power of Self-Supervised Super-Resolution for Resource-Efficient 3D MRI Segmentation
Oct 14, 2024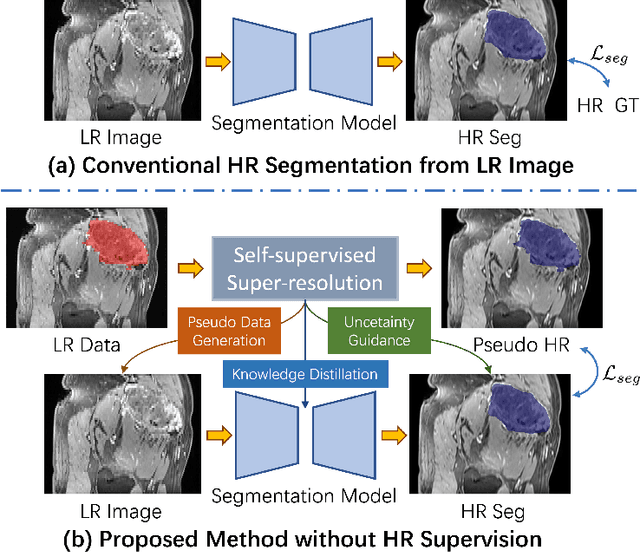
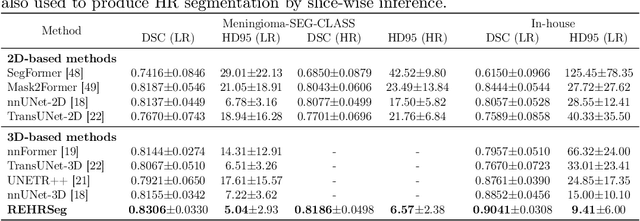
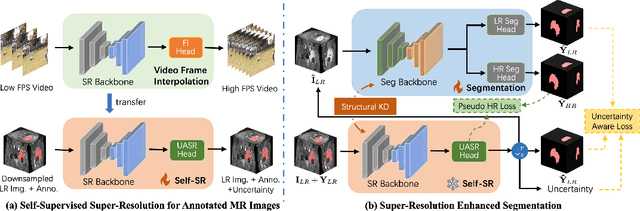
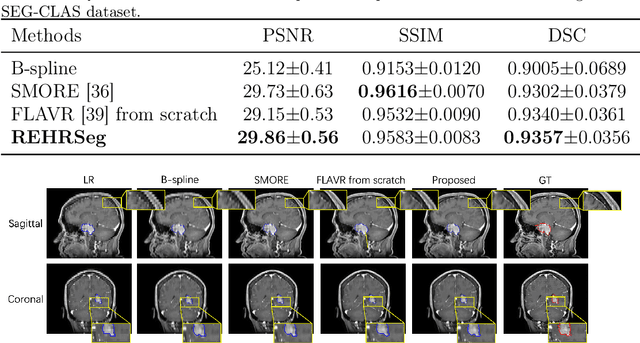
Abstract:High-resolution (HR) 3D magnetic resonance imaging (MRI) can provide detailed anatomical structural information, enabling precise segmentation of regions of interest for various medical image analysis tasks. Due to the high demands of acquisition device, collection of HR images with their annotations is always impractical in clinical scenarios. Consequently, segmentation results based on low-resolution (LR) images with large slice thickness are often unsatisfactory for subsequent tasks. In this paper, we propose a novel Resource-Efficient High-Resolution Segmentation framework (REHRSeg) to address the above-mentioned challenges in real-world applications, which can achieve HR segmentation while only employing the LR images as input. REHRSeg is designed to leverage self-supervised super-resolution (self-SR) to provide pseudo supervision, therefore the relatively easier-to-acquire LR annotated images generated by 2D scanning protocols can be directly used for model training. The main contribution to ensure the effectiveness in self-SR for enhancing segmentation is three-fold: (1) We mitigate the data scarcity problem in the medical field by using pseudo-data for training the segmentation model. (2) We design an uncertainty-aware super-resolution (UASR) head in self-SR to raise the awareness of segmentation uncertainty as commonly appeared on the ROI boundaries. (3) We align the spatial features for self-SR and segmentation through structural knowledge distillation to enable a better capture of region correlations. Experimental results demonstrate that REHRSeg achieves high-quality HR segmentation without intensive supervision, while also significantly improving the baseline performance for LR segmentation.
Inter-slice Super-resolution of Magnetic Resonance Images by Pre-training and Self-supervised Fine-tuning
Jun 10, 2024Abstract:In clinical practice, 2D magnetic resonance (MR) sequences are widely adopted. While individual 2D slices can be stacked to form a 3D volume, the relatively large slice spacing can pose challenges for both image visualization and subsequent analysis tasks, which often require isotropic voxel spacing. To reduce slice spacing, deep-learning-based super-resolution techniques are widely investigated. However, most current solutions require a substantial number of paired high-resolution and low-resolution images for supervised training, which are typically unavailable in real-world scenarios. In this work, we propose a self-supervised super-resolution framework for inter-slice super-resolution of MR images. Our framework is first featured by pre-training on video dataset, as temporal correlation of videos is found beneficial for modeling the spatial relation among MR slices. Then, we use public high-quality MR dataset to fine-tune our pre-trained model, for enhancing awareness of our model to medical data. Finally, given a target dataset at hand, we utilize self-supervised fine-tuning to further ensure our model works well with user-specific super-resolution tasks. The proposed method demonstrates superior performance compared to other self-supervised methods and also holds the potential to benefit various downstream applications.
Two-stage Cytopathological Image Synthesis for Augmenting Cervical Abnormality Screening
Feb 25, 2024Abstract:Automatic thin-prep cytologic test (TCT) screening can assist pathologists in finding cervical abnormality towards accurate and efficient cervical cancer diagnosis. Current automatic TCT screening systems mostly involve abnormal cervical cell detection, which generally requires large-scale and diverse training data with high-quality annotations to achieve promising performance. Pathological image synthesis is naturally raised to minimize the efforts in data collection and annotation. However, it is challenging to generate realistic large-size cytopathological images while simultaneously synthesizing visually plausible appearances for small-size abnormal cervical cells. In this paper, we propose a two-stage image synthesis framework to create synthetic data for augmenting cervical abnormality screening. In the first Global Image Generation stage, a Normal Image Generator is designed to generate cytopathological images full of normal cervical cells. In the second Local Cell Editing stage, normal cells are randomly selected from the generated images and then are converted to different types of abnormal cells using the proposed Abnormal Cell Synthesizer. Both Normal Image Generator and Abnormal Cell Synthesizer are built upon Stable Diffusion, a pre-trained foundation model for image synthesis, via parameter-efficient fine-tuning methods for customizing cytopathological image contents and extending spatial layout controllability, respectively. Our experiments demonstrate the synthetic image quality, diversity, and controllability of the proposed synthesis framework, and validate its data augmentation effectiveness in enhancing the performance of abnormal cervical cell detection.
Uni-COAL: A Unified Framework for Cross-Modality Synthesis and Super-Resolution of MR Images
Nov 14, 2023



Abstract:Cross-modality synthesis (CMS), super-resolution (SR), and their combination (CMSR) have been extensively studied for magnetic resonance imaging (MRI). Their primary goals are to enhance the imaging quality by synthesizing the desired modality and reducing the slice thickness. Despite the promising synthetic results, these techniques are often tailored to specific tasks, thereby limiting their adaptability to complex clinical scenarios. Therefore, it is crucial to build a unified network that can handle various image synthesis tasks with arbitrary requirements of modality and resolution settings, so that the resources for training and deploying the models can be greatly reduced. However, none of the previous works is capable of performing CMS, SR, and CMSR using a unified network. Moreover, these MRI reconstruction methods often treat alias frequencies improperly, resulting in suboptimal detail restoration. In this paper, we propose a Unified Co-Modulated Alias-free framework (Uni-COAL) to accomplish the aforementioned tasks with a single network. The co-modulation design of the image-conditioned and stochastic attribute representations ensures the consistency between CMS and SR, while simultaneously accommodating arbitrary combinations of input/output modalities and thickness. The generator of Uni-COAL is also designed to be alias-free based on the Shannon-Nyquist signal processing framework, ensuring effective suppression of alias frequencies. Additionally, we leverage the semantic prior of Segment Anything Model (SAM) to guide Uni-COAL, ensuring a more authentic preservation of anatomical structures during synthesis. Experiments on three datasets demonstrate that Uni-COAL outperforms the alternatives in CMS, SR, and CMSR tasks for MR images, which highlights its generalizability to wide-range applications.
Progressive Attention Guidance for Whole Slide Vulvovaginal Candidiasis Screening
Sep 06, 2023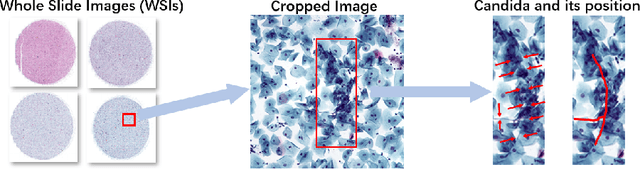
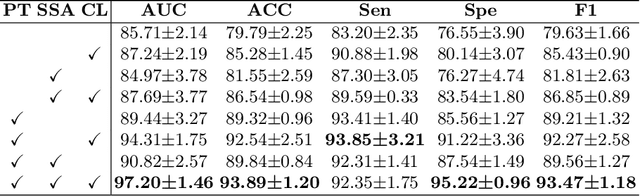
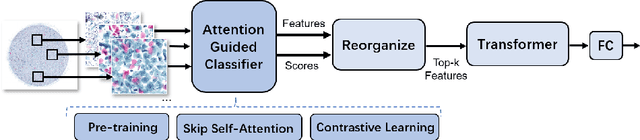
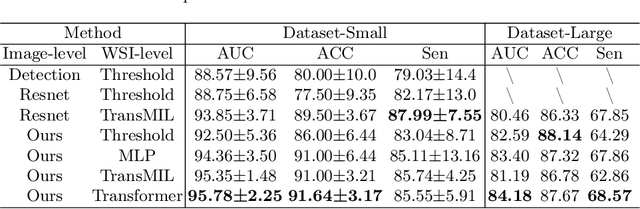
Abstract:Vulvovaginal candidiasis (VVC) is the most prevalent human candidal infection, estimated to afflict approximately 75% of all women at least once in their lifetime. It will lead to several symptoms including pruritus, vaginal soreness, and so on. Automatic whole slide image (WSI) classification is highly demanded, for the huge burden of disease control and prevention. However, the WSI-based computer-aided VCC screening method is still vacant due to the scarce labeled data and unique properties of candida. Candida in WSI is challenging to be captured by conventional classification models due to its distinctive elongated shape, the small proportion of their spatial distribution, and the style gap from WSIs. To make the model focus on the candida easier, we propose an attention-guided method, which can obtain a robust diagnosis classification model. Specifically, we first use a pre-trained detection model as prior instruction to initialize the classification model. Then we design a Skip Self-Attention module to refine the attention onto the fined-grained features of candida. Finally, we use a contrastive learning method to alleviate the overfitting caused by the style gap of WSIs and suppress the attention to false positive regions. Our experimental results demonstrate that our framework achieves state-of-the-art performance. Code and example data are available at https://github.com/cjdbehumble/MICCAI2023-VVC-Screening.
* Accepted in the main conference MICCAI 2023
AdLER: Adversarial Training with Label Error Rectification for One-Shot Medical Image Segmentation
Sep 02, 2023Abstract:Accurate automatic segmentation of medical images typically requires large datasets with high-quality annotations, making it less applicable in clinical settings due to limited training data. One-shot segmentation based on learned transformations (OSSLT) has shown promise when labeled data is extremely limited, typically including unsupervised deformable registration, data augmentation with learned registration, and segmentation learned from augmented data. However, current one-shot segmentation methods are challenged by limited data diversity during augmentation, and potential label errors caused by imperfect registration. To address these issues, we propose a novel one-shot medical image segmentation method with adversarial training and label error rectification (AdLER), with the aim of improving the diversity of generated data and correcting label errors to enhance segmentation performance. Specifically, we implement a novel dual consistency constraint to ensure anatomy-aligned registration that lessens registration errors. Furthermore, we develop an adversarial training strategy to augment the atlas image, which ensures both generation diversity and segmentation robustness. We also propose to rectify potential label errors in the augmented atlas images by estimating segmentation uncertainty, which can compensate for the imperfect nature of deformable registration and improve segmentation authenticity. Experiments on the CANDI and ABIDE datasets demonstrate that the proposed AdLER outperforms previous state-of-the-art methods by 0.7% (CANDI), 3.6% (ABIDE "seen"), and 4.9% (ABIDE "unseen") in segmentation based on Dice scores, respectively. The source code will be available at https://github.com/hsiangyuzhao/AdLER.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge