Kai Xuan
Pathology Image Restoration via Mixture of Prompts
Mar 16, 2025Abstract:In digital pathology, acquiring all-in-focus images is essential to high-quality imaging and high-efficient clinical workflow. Traditional scanners achieve this by scanning at multiple focal planes of varying depths and then merging them, which is relatively slow and often struggles with complex tissue defocus. Recent prevailing image restoration technique provides a means to restore high-quality pathology images from scans of single focal planes. However, existing image restoration methods are inadequate, due to intricate defocus patterns in pathology images and their domain-specific semantic complexities. In this work, we devise a two-stage restoration solution cascading a transformer and a diffusion model, to benefit from their powers in preserving image fidelity and perceptual quality, respectively. We particularly propose a novel mixture of prompts for the two-stage solution. Given initial prompt that models defocus in microscopic imaging, we design two prompts that describe the high-level image semantics from pathology foundation model and the fine-grained tissue structures via edge extraction. We demonstrate that, by feeding the prompt mixture to our method, we can restore high-quality pathology images from single-focal-plane scans, implying high potentials of the mixture of prompts to clinical usage. Code will be publicly available at https://github.com/caijd2000/MoP.
Arbitrary Reduction of MRI Inter-slice Spacing Using Hierarchical Feature Conditional Diffusion
Apr 18, 2023
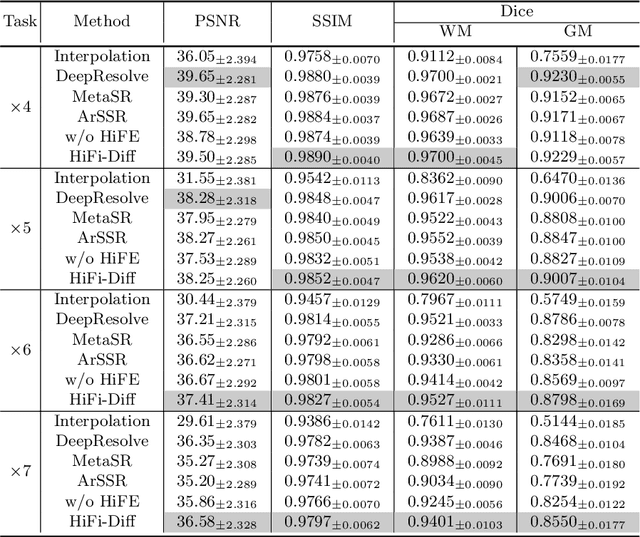


Abstract:Magnetic resonance (MR) images collected in 2D scanning protocols typically have large inter-slice spacing, resulting in high in-plane resolution but reduced through-plane resolution. Super-resolution techniques can reduce the inter-slice spacing of 2D scanned MR images, facilitating the downstream visual experience and computer-aided diagnosis. However, most existing super-resolution methods are trained at a fixed scaling ratio, which is inconvenient in clinical settings where MR scanning may have varying inter-slice spacings. To solve this issue, we propose Hierarchical Feature Conditional Diffusion (HiFi-Diff)} for arbitrary reduction of MR inter-slice spacing. Given two adjacent MR slices and the relative positional offset, HiFi-Diff can iteratively convert a Gaussian noise map into any desired in-between MR slice. Furthermore, to enable fine-grained conditioning, the Hierarchical Feature Extraction (HiFE) module is proposed to hierarchically extract conditional features and conduct element-wise modulation. Our experimental results on the publicly available HCP-1200 dataset demonstrate the high-fidelity super-resolution capability of HiFi-Diff and its efficacy in enhancing downstream segmentation performance.
TBI-GAN: An Adversarial Learning Approach for Data Synthesis on Traumatic Brain Segmentation
Aug 12, 2022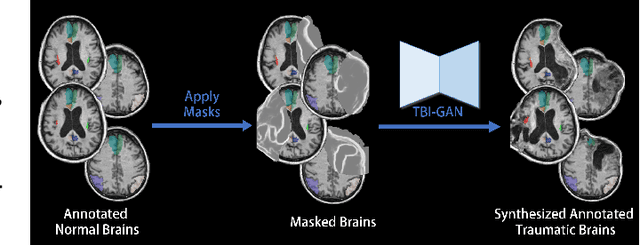
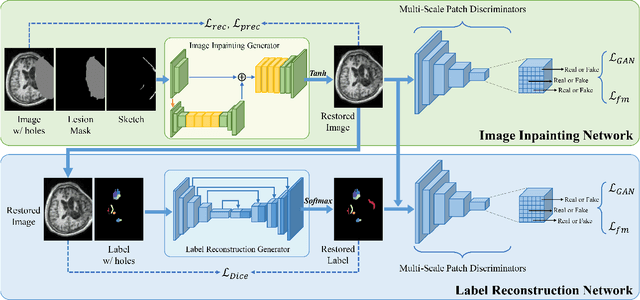
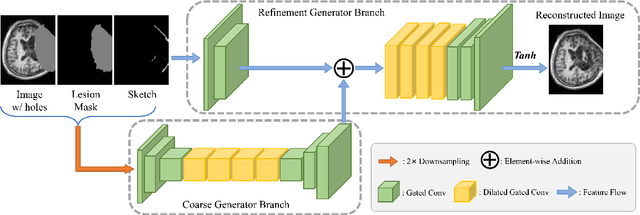
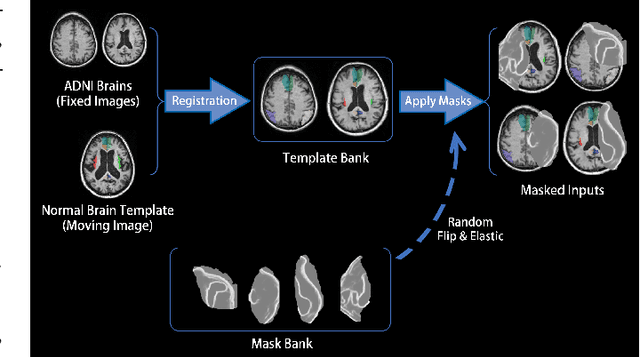
Abstract:Brain network analysis for traumatic brain injury (TBI) patients is critical for its consciousness level assessment and prognosis evaluation, which requires the segmentation of certain consciousness-related brain regions. However, it is difficult to construct a TBI segmentation model as manually annotated MR scans of TBI patients are hard to collect. Data augmentation techniques can be applied to alleviate the issue of data scarcity. However, conventional data augmentation strategies such as spatial and intensity transformation are unable to mimic the deformation and lesions in traumatic brains, which limits the performance of the subsequent segmentation task. To address these issues, we propose a novel medical image inpainting model named TBI-GAN to synthesize TBI MR scans with paired brain label maps. The main strength of our TBI-GAN method is that it can generate TBI images and corresponding label maps simultaneously, which has not been achieved in the previous inpainting methods for medical images. We first generate the inpainted image under the guidance of edge information following a coarse-to-fine manner, and then the synthesized intensity image is used as the prior for label inpainting. Furthermore, we introduce a registration-based template augmentation pipeline to increase the diversity of the synthesized image pairs and enhance the capacity of data augmentation. Experimental results show that the proposed TBI-GAN method can produce sufficient synthesized TBI images with high quality and valid label maps, which can greatly improve the 2D and 3D traumatic brain segmentation performance compared with the alternatives.
Arbitrary Reduction of MRI Slice Spacing Based on Local-Aware Implicit Representation
May 23, 2022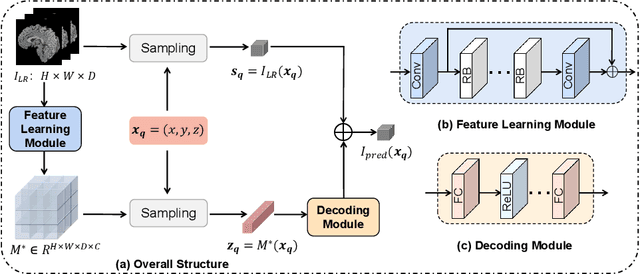
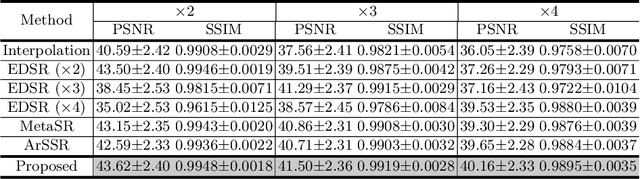

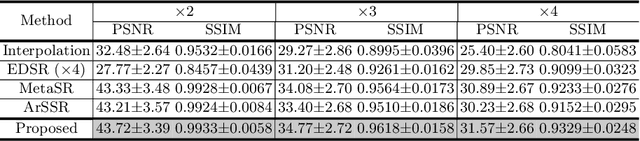
Abstract:Magnetic resonance (MR) images are often acquired in 2D settings for real clinical applications. The 3D volumes reconstructed by stacking multiple 2D slices have large inter-slice spacing, resulting in lower inter-slice resolution than intra-slice resolution. Super-resolution is a powerful tool to reduce the inter-slice spacing of 3D images to facilitate subsequent visualization and computation tasks. However, most existing works train the super-resolution network at a fixed ratio, which is inconvenient in clinical scenes due to the heterogeneous parameters in MR scanning. In this paper, we propose a single super-resolution network to reduce the inter-slice spacing of MR images at an arbitrarily adjustable ratio. Specifically, we view the input image as a continuous implicit function of coordinates. The intermediate slices of different spacing ratios could be constructed according to the implicit representation up-sampled in the continuous domain. We particularly propose a novel local-aware spatial attention mechanism and long-range residual learning to boost the quality of the output image. The experimental results demonstrate the superiority of our proposed method, even compared to the models trained at a fixed ratio.
Knee Cartilage Defect Assessment by Graph Representation and Surface Convolution
Jan 12, 2022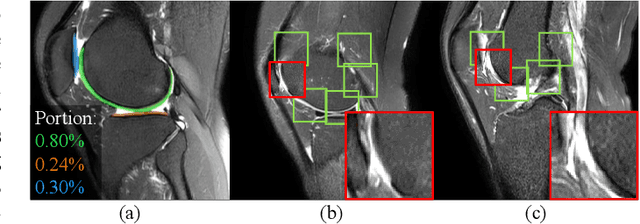
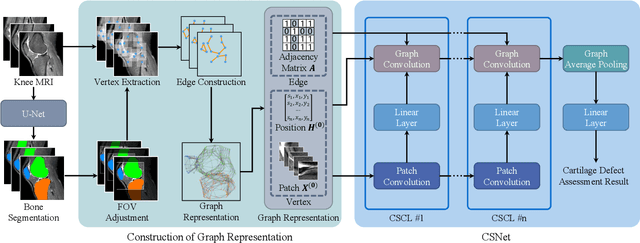
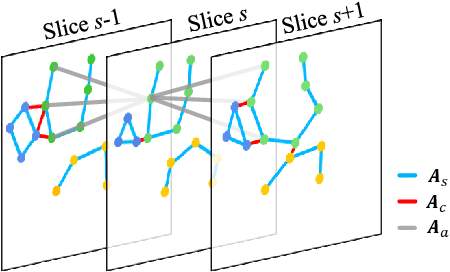
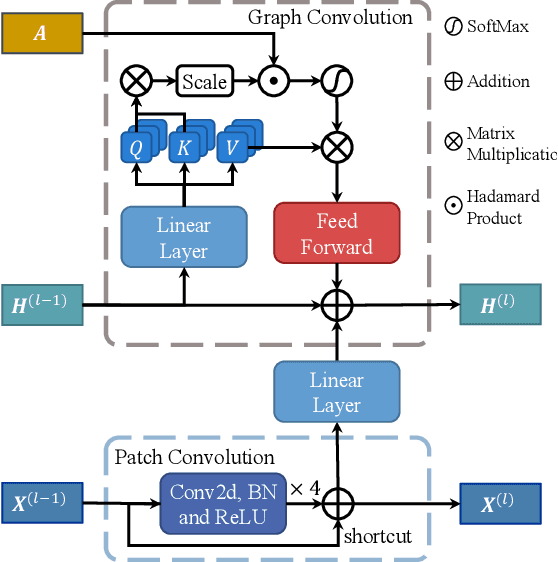
Abstract:Knee osteoarthritis (OA) is the most common osteoarthritis and a leading cause of disability. Cartilage defects are regarded as major manifestations of knee OA, which are visible by magnetic resonance imaging (MRI). Thus early detection and assessment for knee cartilage defects are important for protecting patients from knee OA. In this way, many attempts have been made on knee cartilage defect assessment by applying convolutional neural networks (CNNs) to knee MRI. However, the physiologic characteristics of the cartilage may hinder such efforts: the cartilage is a thin curved layer, implying that only a small portion of voxels in knee MRI can contribute to the cartilage defect assessment; heterogeneous scanning protocols further challenge the feasibility of the CNNs in clinical practice; the CNN-based knee cartilage evaluation results lack interpretability. To address these challenges, we model the cartilages structure and appearance from knee MRI into a graph representation, which is capable of handling highly diverse clinical data. Then, guided by the cartilage graph representation, we design a non-Euclidean deep learning network with the self-attention mechanism, to extract cartilage features in the local and global, and to derive the final assessment with a visualized result. Our comprehensive experiments show that the proposed method yields superior performance in knee cartilage defect assessment, plus its convenient 3D visualization for interpretability.
Multi-Modal MRI Reconstruction Assisted with Spatial Alignment Network
Aug 30, 2021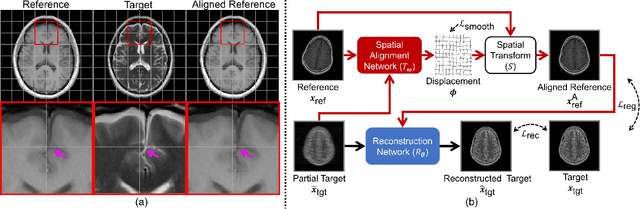
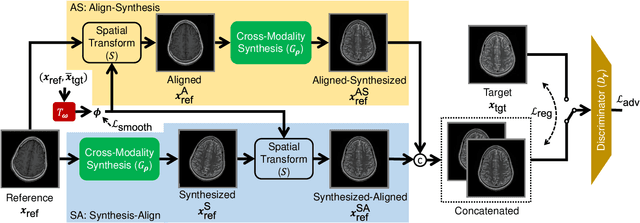
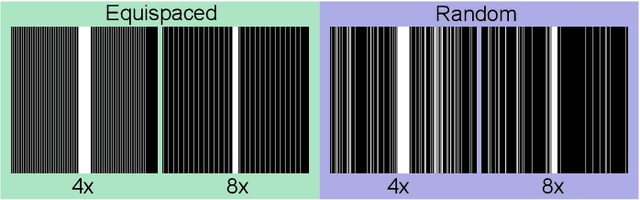
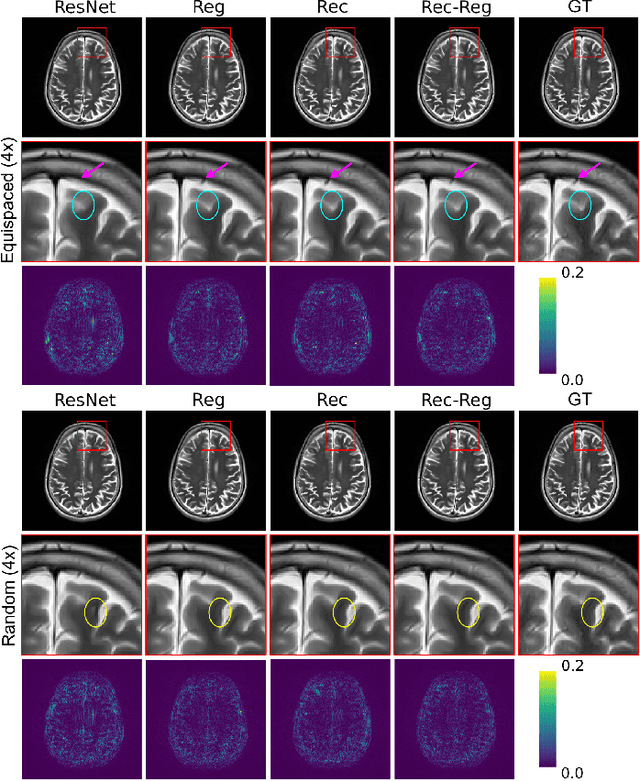
Abstract:In clinical practice, magnetic resonance imaging (MRI) with multiple contrasts is usually acquired in a single study to assess different properties of the same region of interest in human body. The whole acquisition process can be accelerated by having one or more modalities under-sampled in the $k$-space. Recent researches demonstrate that, considering the redundancy between different contrasts or modalities, a target MRI modality under-sampled in the $k$-space can be more efficiently reconstructed with a fully-sampled MRI contrast as the reference modality. However, we find that the performance of the above multi-modal reconstruction can be negatively affected by subtle spatial misalignment between different contrasts, which is actually common in clinical practice. In this paper, to compensate for such spatial misalignment, we integrate the spatial alignment network with multi-modal reconstruction towards better reconstruction quality of the target modality. First, the spatial alignment network estimates the spatial misalignment between the fully-sampled reference and the under-sampled target images, and warps the reference image accordingly. Then, the aligned fully-sampled reference image joins the multi-modal reconstruction of the under-sampled target image. Also, considering the contrast difference between the target and the reference images, we particularly design the cross-modality-synthesis-based registration loss, in combination with the reconstruction loss, to jointly train the spatial alignment network and the reconstruction network. Experiments on both clinical MRI and multi-coil $k$-space raw data demonstrate the superiority and robustness of multi-modal MRI reconstruction empowered with our spatial alignment network. Our code is publicly available at \url{https://github.com/woxuankai/SpatialAlignmentNetwork}.
Generalized-TODIM Method for Multi-criteria Decision Making with Basic Uncertain Information and its Application
Apr 27, 2021
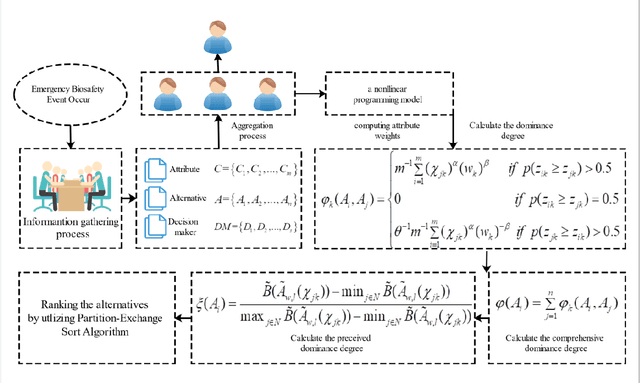
Abstract:Due to the fact that basic uncertain information provides a simple form for decision information with certainty degree, it has been developed to reflect the quality of observed or subjective assessments. In order to study the algebra structure and preference relation of basic uncertain information, we develop some algebra operations for basic uncertain information. The order relation of such type of information has also been considered. Finally, to apply the developed algebra operations and order relations, a generalized TODIM method for multi-attribute decision making with basic uncertain information is given. The numerical example shows that the developed decision procedure is valid.
A Self-ensembling Framework for Semi-supervised Knee Osteoarthritis Localization and Classification with Dual-Consistency
May 19, 2020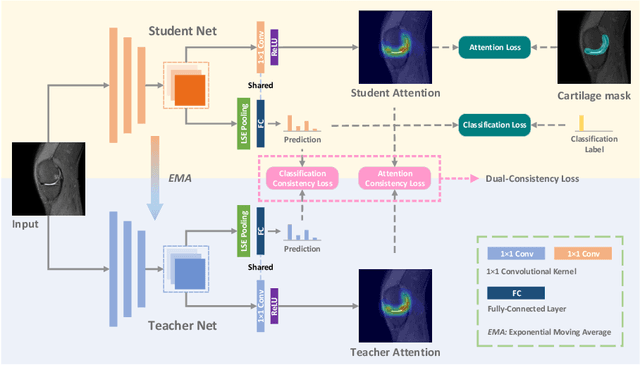
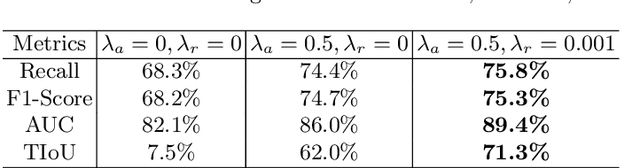
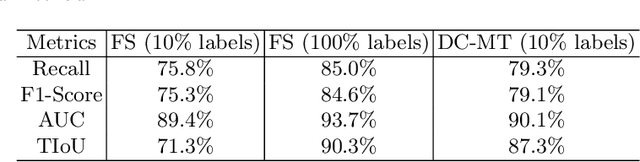
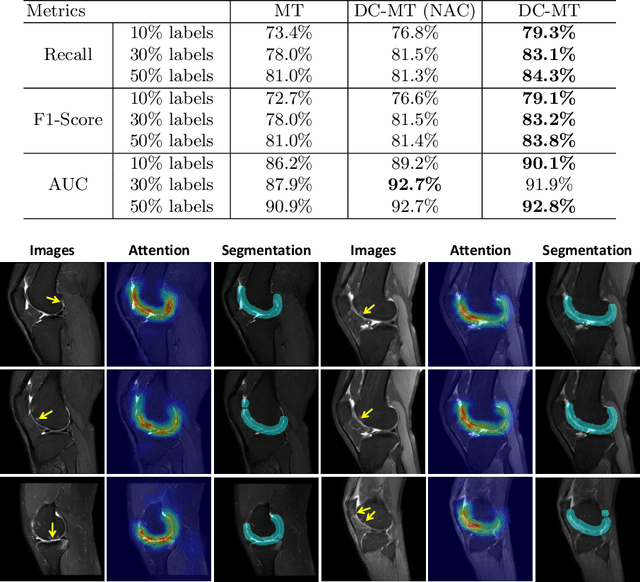
Abstract:Knee osteoarthritis (OA) is one of the most common musculoskeletal disorders and requires early-stage diagnosis. Nowadays, the deep convolutional neural networks have achieved greatly in the computer-aided diagnosis field. However, the construction of the deep learning models usually requires great amounts of annotated data, which is generally high-cost. In this paper, we propose a novel approach for knee OA diagnosis, including severity classification and lesion localization. Particularly, we design a self-ensembling framework, which is composed of a student network and a teacher network with the same structure. The student network learns from both labeled data and unlabeled data and the teacher network averages the student model weights through the training course. A novel attention loss function is developed to obtain accurate attention masks. With dual-consistency checking of the attention in the lesion classification and localization, the two networks can gradually optimize the attention distribution and improve the performance of each other, whereas the training relies on partially labeled data only and follows the semi-supervised manner. Experiments show that the proposed method can significantly improve the self-ensembling performance in both knee OA classification and localization, and also greatly reduce the needs of annotated data.
Robust Brain Magnetic Resonance Image Segmentation for Hydrocephalus Patients: Hard and Soft Attention
Jan 12, 2020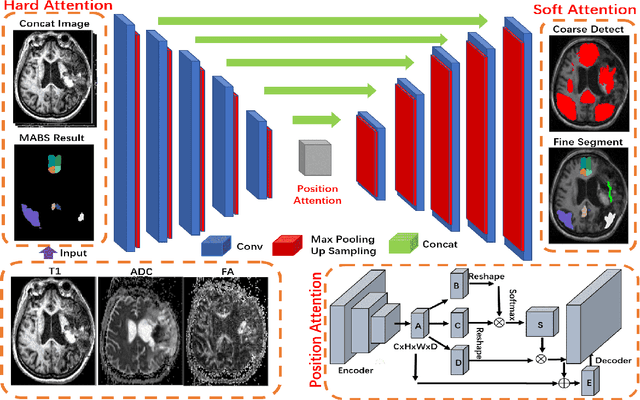
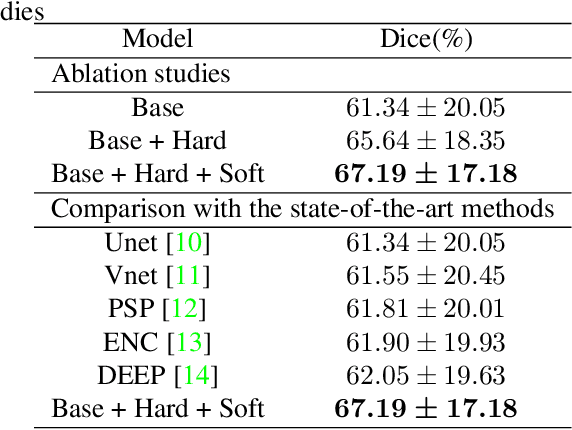
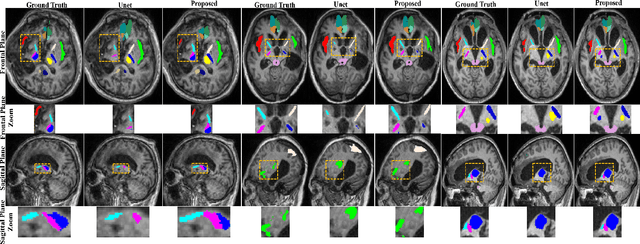
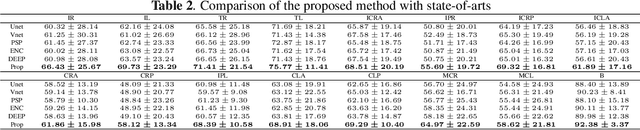
Abstract:Brain magnetic resonance (MR) segmentation for hydrocephalus patients is considered as a challenging work. Encoding the variation of the brain anatomical structures from different individuals cannot be easily achieved. The task becomes even more difficult especially when the image data from hydrocephalus patients are considered, which often have large deformations and differ significantly from the normal subjects. Here, we propose a novel strategy with hard and soft attention modules to solve the segmentation problems for hydrocephalus MR images. Our main contributions are three-fold: 1) the hard-attention module generates coarse segmentation map using multi-atlas-based method and the VoxelMorph tool, which guides subsequent segmentation process and improves its robustness; 2) the soft-attention module incorporates position attention to capture precise context information, which further improves the segmentation accuracy; 3) we validate our method by segmenting insula, thalamus and many other regions-of-interests (ROIs) that are critical to quantify brain MR images of hydrocephalus patients in real clinical scenario. The proposed method achieves much improved robustness and accuracy when segmenting all 17 consciousness-related ROIs with high variations for different subjects. To the best of our knowledge, this is the first work to employ deep learning for solving the brain segmentation problems of hydrocephalus patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge