Lei Xiang
CT-MVSNet: Efficient Multi-View Stereo with Cross-scale Transformer
Dec 14, 2023Abstract:Recent deep multi-view stereo (MVS) methods have widely incorporated transformers into cascade network for high-resolution depth estimation, achieving impressive results. However, existing transformer-based methods are constrained by their computational costs, preventing their extension to finer stages. In this paper, we propose a novel cross-scale transformer (CT) that processes feature representations at different stages without additional computation. Specifically, we introduce an adaptive matching-aware transformer (AMT) that employs different interactive attention combinations at multiple scales. This combined strategy enables our network to capture intra-image context information and enhance inter-image feature relationships. Besides, we present a dual-feature guided aggregation (DFGA) that embeds the coarse global semantic information into the finer cost volume construction to further strengthen global and local feature awareness. Meanwhile, we design a feature metric loss (FM Loss) that evaluates the feature bias before and after transformation to reduce the impact of feature mismatch on depth estimation. Extensive experiments on DTU dataset and Tanks and Temples (T\&T) benchmark demonstrate that our method achieves state-of-the-art results. Code is available at https://github.com/wscstrive/CT-MVSNet.
MD-IQA: Learning Multi-scale Distributed Image Quality Assessment with Semi Supervised Learning for Low Dose CT
Nov 14, 2023Abstract:Image quality assessment (IQA) plays a critical role in optimizing radiation dose and developing novel medical imaging techniques in computed tomography (CT). Traditional IQA methods relying on hand-crafted features have limitations in summarizing the subjective perceptual experience of image quality. Recent deep learning-based approaches have demonstrated strong modeling capabilities and potential for medical IQA, but challenges remain regarding model generalization and perceptual accuracy. In this work, we propose a multi-scale distributions regression approach to predict quality scores by constraining the output distribution, thereby improving model generalization. Furthermore, we design a dual-branch alignment network to enhance feature extraction capabilities. Additionally, semi-supervised learning is introduced by utilizing pseudo-labels for unlabeled data to guide model training. Extensive qualitative experiments demonstrate the effectiveness of our proposed method for advancing the state-of-the-art in deep learning-based medical IQA. Code is available at: https://github.com/zunzhumu/MD-IQA.
Dual Feature Augmentation Network for Generalized Zero-shot Learning
Sep 25, 2023Abstract:Zero-shot learning (ZSL) aims to infer novel classes without training samples by transferring knowledge from seen classes. Existing embedding-based approaches for ZSL typically employ attention mechanisms to locate attributes on an image. However, these methods often ignore the complex entanglement among different attributes' visual features in the embedding space. Additionally, these methods employ a direct attribute prediction scheme for classification, which does not account for the diversity of attributes in images of the same category. To address these issues, we propose a novel Dual Feature Augmentation Network (DFAN), which comprises two feature augmentation modules, one for visual features and the other for semantic features. The visual feature augmentation module explicitly learns attribute features and employs cosine distance to separate them, thus enhancing attribute representation. In the semantic feature augmentation module, we propose a bias learner to capture the offset that bridges the gap between actual and predicted attribute values from a dataset's perspective. Furthermore, we introduce two predictors to reconcile the conflicts between local and global features. Experimental results on three benchmarks demonstrate the marked advancement of our method compared to state-of-the-art approaches. Our code is available at https://github.com/Sion1/DFAN.
Whole-body tumor segmentation of 18F -FDG PET/CT using a cascaded and ensembled convolutional neural networks
Oct 14, 2022



Abstract:Background: A crucial initial processing step for quantitative PET/CT analysis is the segmentation of tumor lesions enabling accurate feature ex-traction, tumor characterization, oncologic staging, and image-based therapy response assessment. Manual lesion segmentation is however associated with enormous effort and cost and is thus infeasible in clinical routine. Goal: The goal of this study was to report the performance of a deep neural network designed to automatically segment regions suspected of cancer in whole-body 18F-FDG PET/CT images in the context of the AutoPET challenge. Method: A cascaded approach was developed where a stacked ensemble of 3D UNET CNN processed the PET/CT images at a fixed 6mm resolution. A refiner network composed of residual layers enhanced the 6mm segmentation mask to the original resolution. Results: 930 cases were used to train the model. 50% were histologically proven cancer patients and 50% were healthy controls. We obtained a dice=0.68 on 84 stratified test cases. Manual and automatic Metabolic Tumor Volume (MTV) were highly correlated (R2 = 0.969,Slope = 0.947). Inference time was 89.7 seconds on average. Conclusion: The proposed algorithm accurately segmented regions suspicious for cancer in whole-body 18F -FDG PET/CT images.
Multi-Modal MRI Reconstruction Assisted with Spatial Alignment Network
Aug 30, 2021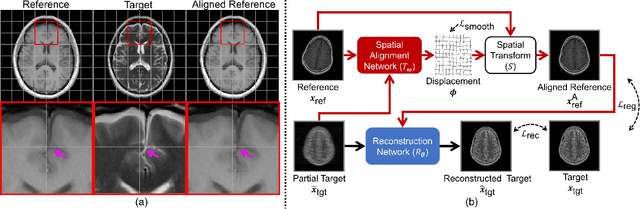
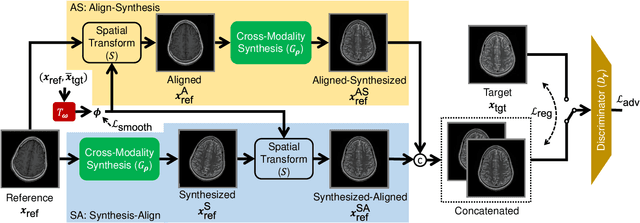
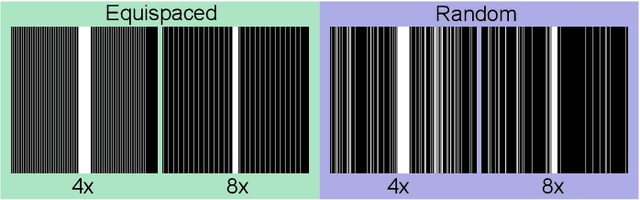
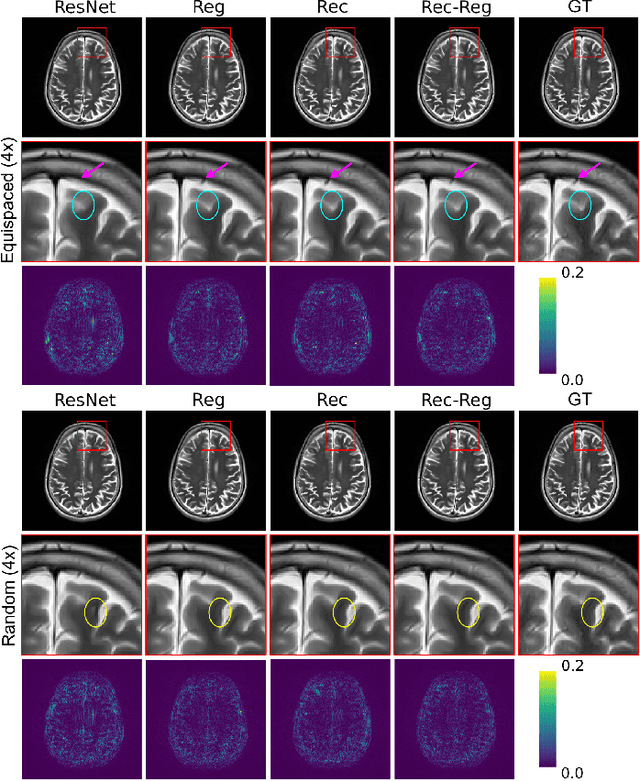
Abstract:In clinical practice, magnetic resonance imaging (MRI) with multiple contrasts is usually acquired in a single study to assess different properties of the same region of interest in human body. The whole acquisition process can be accelerated by having one or more modalities under-sampled in the $k$-space. Recent researches demonstrate that, considering the redundancy between different contrasts or modalities, a target MRI modality under-sampled in the $k$-space can be more efficiently reconstructed with a fully-sampled MRI contrast as the reference modality. However, we find that the performance of the above multi-modal reconstruction can be negatively affected by subtle spatial misalignment between different contrasts, which is actually common in clinical practice. In this paper, to compensate for such spatial misalignment, we integrate the spatial alignment network with multi-modal reconstruction towards better reconstruction quality of the target modality. First, the spatial alignment network estimates the spatial misalignment between the fully-sampled reference and the under-sampled target images, and warps the reference image accordingly. Then, the aligned fully-sampled reference image joins the multi-modal reconstruction of the under-sampled target image. Also, considering the contrast difference between the target and the reference images, we particularly design the cross-modality-synthesis-based registration loss, in combination with the reconstruction loss, to jointly train the spatial alignment network and the reconstruction network. Experiments on both clinical MRI and multi-coil $k$-space raw data demonstrate the superiority and robustness of multi-modal MRI reconstruction empowered with our spatial alignment network. Our code is publicly available at \url{https://github.com/woxuankai/SpatialAlignmentNetwork}.
Multi-Scale Boosted Dehazing Network with Dense Feature Fusion
Apr 28, 2020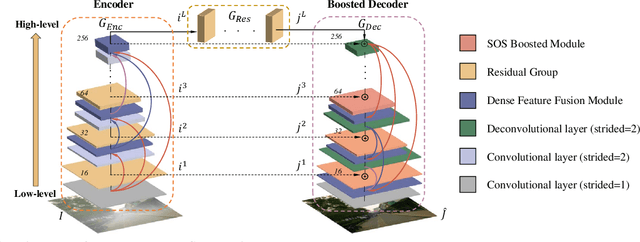
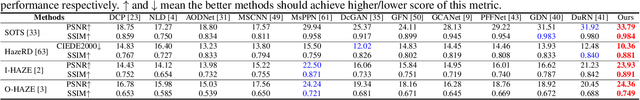
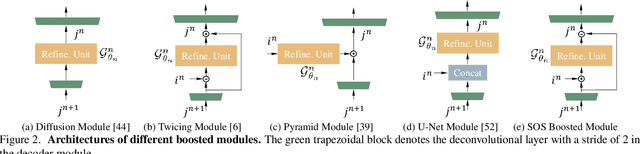
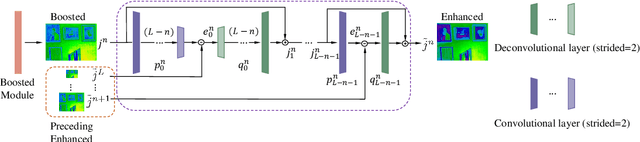
Abstract:In this paper, we propose a Multi-Scale Boosted Dehazing Network with Dense Feature Fusion based on the U-Net architecture. The proposed method is designed based on two principles, boosting and error feedback, and we show that they are suitable for the dehazing problem. By incorporating the Strengthen-Operate-Subtract boosting strategy in the decoder of the proposed model, we develop a simple yet effective boosted decoder to progressively restore the haze-free image. To address the issue of preserving spatial information in the U-Net architecture, we design a dense feature fusion module using the back-projection feedback scheme. We show that the dense feature fusion module can simultaneously remedy the missing spatial information from high-resolution features and exploit the non-adjacent features. Extensive evaluations demonstrate that the proposed model performs favorably against the state-of-the-art approaches on the benchmark datasets as well as real-world hazy images.
Dual Adversarial Learning with Attention Mechanism for Fine-grained Medical Image Synthesis
Jul 07, 2019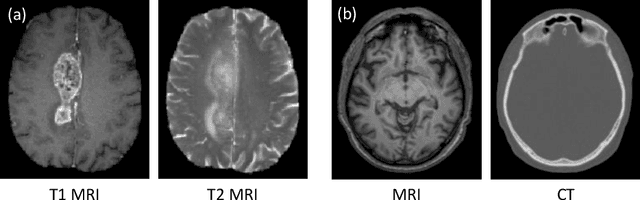
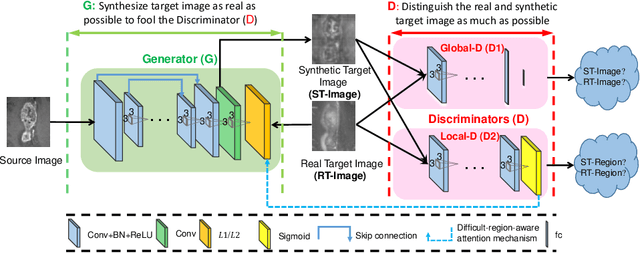

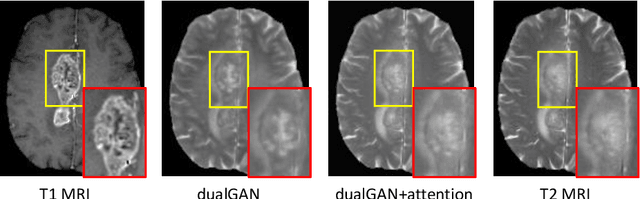
Abstract:Medical imaging plays a critical role in various clinical applications. However, due to multiple considerations such as cost and risk, the acquisition of certain image modalities could be limited. To address this issue, many cross-modality medical image synthesis methods have been proposed. However, the current methods cannot well model the hard-to-synthesis regions (e.g., tumor or lesion regions). To address this issue, we propose a simple but effective strategy, that is, we propose a dual-discriminator (dual-D) adversarial learning system, in which, a global-D is used to make an overall evaluation for the synthetic image, and a local-D is proposed to densely evaluate the local regions of the synthetic image. More importantly, we build an adversarial attention mechanism which targets at better modeling hard-to-synthesize regions (e.g., tumor or lesion regions) based on the local-D. Experimental results show the robustness and accuracy of our method in synthesizing fine-grained target images from the corresponding source images. In particular, we evaluate our method on two datasets, i.e., to address the tasks of generating T2 MRI from T1 MRI for the brain tumor images and generating MRI from CT. Our method outperforms the state-of-the-art methods under comparison in all datasets and tasks. And the proposed difficult-region-aware attention mechanism is also proved to be able to help generate more realistic images, especially for the hard-to-synthesize regions.
Task Decomposition and Synchronization for Semantic Biomedical Image Segmentation
May 21, 2019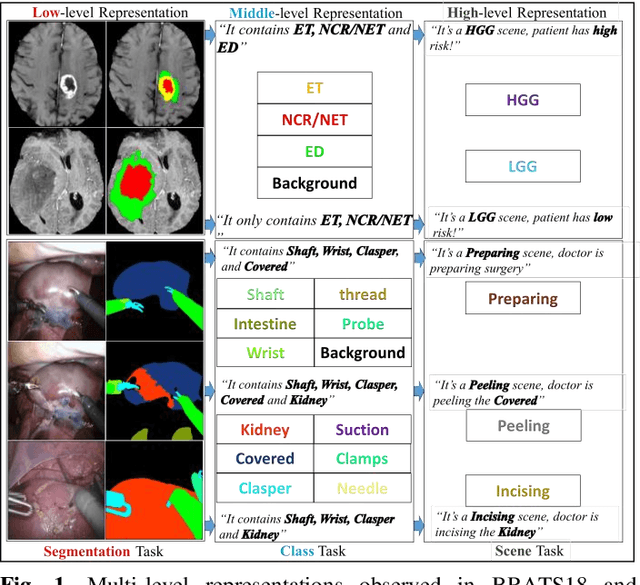
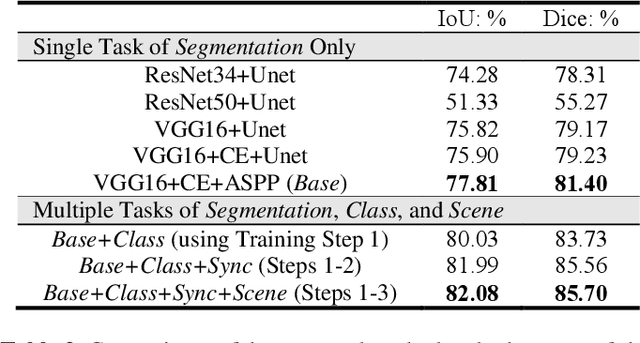
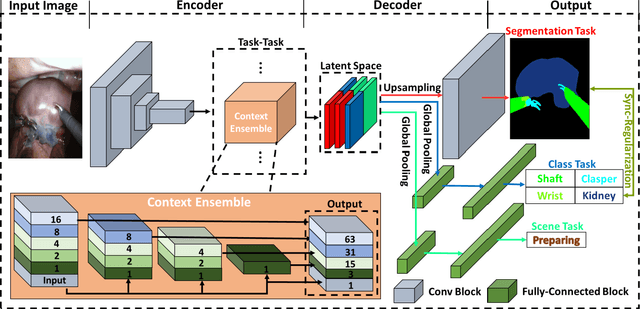
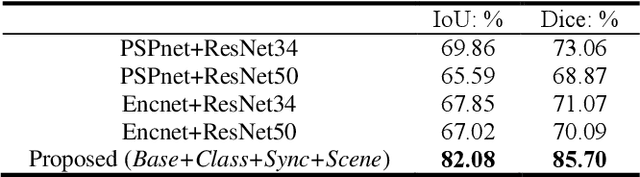
Abstract:Semantic segmentation is essentially important to biomedical image analysis. Many recent works mainly focus on integrating the Fully Convolutional Network (FCN) architecture with sophisticated convolution implementation and deep supervision. In this paper, we propose to decompose the single segmentation task into three subsequent sub-tasks, including (1) pixel-wise image segmentation, (2) prediction of the class labels of the objects within the image, and (3) classification of the scene the image belonging to. While these three sub-tasks are trained to optimize their individual loss functions of different perceptual levels, we propose to let them interact by the task-task context ensemble. Moreover, we propose a novel sync-regularization to penalize the deviation between the outputs of the pixel-wise segmentation and the class prediction tasks. These effective regularizations help FCN utilize context information comprehensively and attain accurate semantic segmentation, even though the number of the images for training may be limited in many biomedical applications. We have successfully applied our framework to three diverse 2D/3D medical image datasets, including Robotic Scene Segmentation Challenge 18 (ROBOT18), Brain Tumor Segmentation Challenge 18 (BRATS18), and Retinal Fundus Glaucoma Challenge (REFUGE18). We have achieved top-tier performance in all three challenges.
Deep Embedding Convolutional Neural Network for Synthesizing CT Image from T1-Weighted MR Image
Nov 09, 2017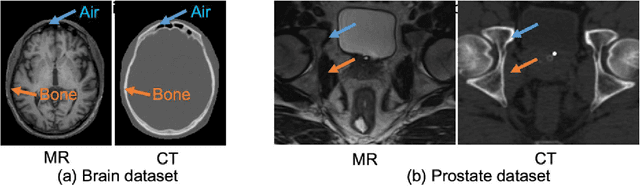
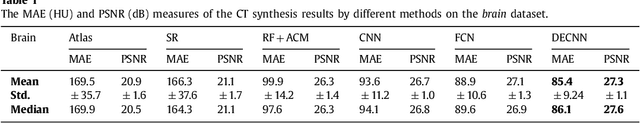
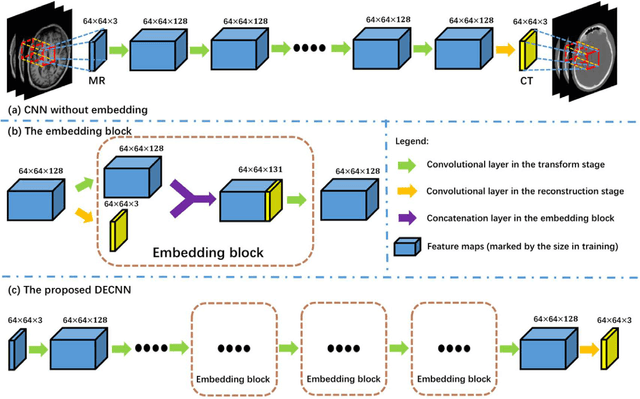
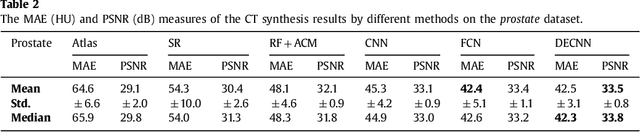
Abstract:Recently, more and more attention is drawn to the field of medical image synthesis across modalities. Among them, the synthesis of computed tomography (CT) image from T1-weighted magnetic resonance (MR) image is of great importance, although the mapping between them is highly complex due to large gaps of appearances of the two modalities. In this work, we aim to tackle this MR-to-CT synthesis by a novel deep embedding convolutional neural network (DECNN). Specifically, we generate the feature maps from MR images, and then transform these feature maps forward through convolutional layers in the network. We can further compute a tentative CT synthesis from the midway of the flow of feature maps, and then embed this tentative CT synthesis back to the feature maps. This embedding operation results in better feature maps, which are further transformed forward in DECNN. After repeat-ing this embedding procedure for several times in the network, we can eventually synthesize a final CT image in the end of the DECNN. We have validated our proposed method on both brain and prostate datasets, by also compar-ing with the state-of-the-art methods. Experimental results suggest that our DECNN (with repeated embedding op-erations) demonstrates its superior performances, in terms of both the perceptive quality of the synthesized CT image and the run-time cost for synthesizing a CT image.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge