Di Zang
Uni-COAL: A Unified Framework for Cross-Modality Synthesis and Super-Resolution of MR Images
Nov 14, 2023



Abstract:Cross-modality synthesis (CMS), super-resolution (SR), and their combination (CMSR) have been extensively studied for magnetic resonance imaging (MRI). Their primary goals are to enhance the imaging quality by synthesizing the desired modality and reducing the slice thickness. Despite the promising synthetic results, these techniques are often tailored to specific tasks, thereby limiting their adaptability to complex clinical scenarios. Therefore, it is crucial to build a unified network that can handle various image synthesis tasks with arbitrary requirements of modality and resolution settings, so that the resources for training and deploying the models can be greatly reduced. However, none of the previous works is capable of performing CMS, SR, and CMSR using a unified network. Moreover, these MRI reconstruction methods often treat alias frequencies improperly, resulting in suboptimal detail restoration. In this paper, we propose a Unified Co-Modulated Alias-free framework (Uni-COAL) to accomplish the aforementioned tasks with a single network. The co-modulation design of the image-conditioned and stochastic attribute representations ensures the consistency between CMS and SR, while simultaneously accommodating arbitrary combinations of input/output modalities and thickness. The generator of Uni-COAL is also designed to be alias-free based on the Shannon-Nyquist signal processing framework, ensuring effective suppression of alias frequencies. Additionally, we leverage the semantic prior of Segment Anything Model (SAM) to guide Uni-COAL, ensuring a more authentic preservation of anatomical structures during synthesis. Experiments on three datasets demonstrate that Uni-COAL outperforms the alternatives in CMS, SR, and CMSR tasks for MR images, which highlights its generalizability to wide-range applications.
TBI-GAN: An Adversarial Learning Approach for Data Synthesis on Traumatic Brain Segmentation
Aug 12, 2022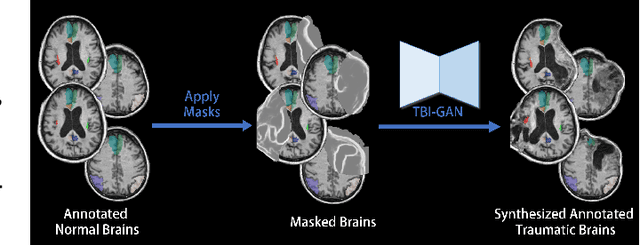
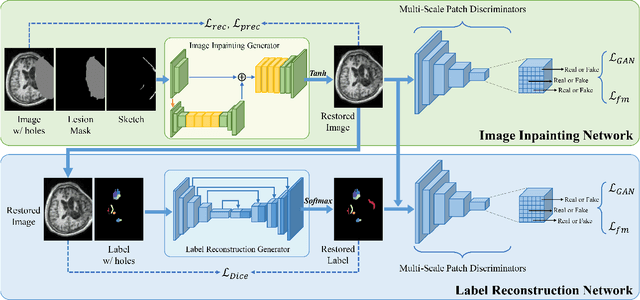
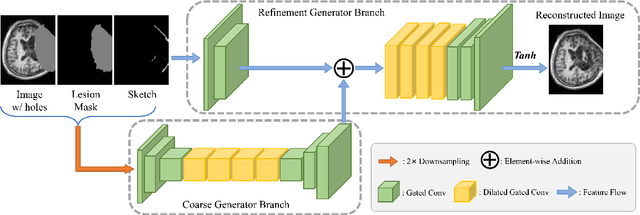
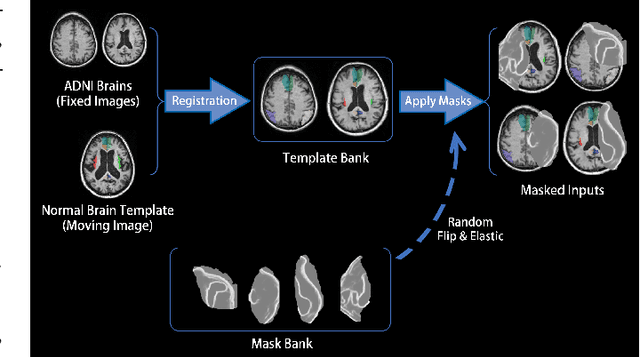
Abstract:Brain network analysis for traumatic brain injury (TBI) patients is critical for its consciousness level assessment and prognosis evaluation, which requires the segmentation of certain consciousness-related brain regions. However, it is difficult to construct a TBI segmentation model as manually annotated MR scans of TBI patients are hard to collect. Data augmentation techniques can be applied to alleviate the issue of data scarcity. However, conventional data augmentation strategies such as spatial and intensity transformation are unable to mimic the deformation and lesions in traumatic brains, which limits the performance of the subsequent segmentation task. To address these issues, we propose a novel medical image inpainting model named TBI-GAN to synthesize TBI MR scans with paired brain label maps. The main strength of our TBI-GAN method is that it can generate TBI images and corresponding label maps simultaneously, which has not been achieved in the previous inpainting methods for medical images. We first generate the inpainted image under the guidance of edge information following a coarse-to-fine manner, and then the synthesized intensity image is used as the prior for label inpainting. Furthermore, we introduce a registration-based template augmentation pipeline to increase the diversity of the synthesized image pairs and enhance the capacity of data augmentation. Experimental results show that the proposed TBI-GAN method can produce sufficient synthesized TBI images with high quality and valid label maps, which can greatly improve the 2D and 3D traumatic brain segmentation performance compared with the alternatives.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge