Chunfeng Lian
FEAT: A Multi-Agent Forensic AI System with Domain-Adapted Large Language Model for Automated Cause-of-Death Analysis
Aug 11, 2025Abstract:Forensic cause-of-death determination faces systemic challenges, including workforce shortages and diagnostic variability, particularly in high-volume systems like China's medicolegal infrastructure. We introduce FEAT (ForEnsic AgenT), a multi-agent AI framework that automates and standardizes death investigations through a domain-adapted large language model. FEAT's application-oriented architecture integrates: (i) a central Planner for task decomposition, (ii) specialized Local Solvers for evidence analysis, (iii) a Memory & Reflection module for iterative refinement, and (iv) a Global Solver for conclusion synthesis. The system employs tool-augmented reasoning, hierarchical retrieval-augmented generation, forensic-tuned LLMs, and human-in-the-loop feedback to ensure legal and medical validity. In evaluations across diverse Chinese case cohorts, FEAT outperformed state-of-the-art AI systems in both long-form autopsy analyses and concise cause-of-death conclusions. It demonstrated robust generalization across six geographic regions and achieved high expert concordance in blinded validations. Senior pathologists validated FEAT's outputs as comparable to those of human experts, with improved detection of subtle evidentiary nuances. To our knowledge, FEAT is the first LLM-based AI agent system dedicated to forensic medicine, offering scalable, consistent death certification while maintaining expert-level rigor. By integrating AI efficiency with human oversight, this work could advance equitable access to reliable medicolegal services while addressing critical capacity constraints in forensic systems.
R2Gen-Mamba: A Selective State Space Model for Radiology Report Generation
Oct 21, 2024Abstract:Radiology report generation is crucial in medical imaging,but the manual annotation process by physicians is time-consuming and labor-intensive, necessitating the develop-ment of automatic report generation methods. Existingresearch predominantly utilizes Transformers to generateradiology reports, which can be computationally intensive,limiting their use in real applications. In this work, we presentR2Gen-Mamba, a novel automatic radiology report genera-tion method that leverages the efficient sequence processingof the Mamba with the contextual benefits of Transformerarchitectures. Due to lower computational complexity ofMamba, R2Gen-Mamba not only enhances training and in-ference efficiency but also produces high-quality reports.Experimental results on two benchmark datasets with morethan 210,000 X-ray image-report pairs demonstrate the ef-fectiveness of R2Gen-Mamba regarding report quality andcomputational efficiency compared with several state-of-the-art methods. The source code can be accessed online.
Large-vocabulary forensic pathological analyses via prototypical cross-modal contrastive learning
Jul 20, 2024Abstract:Forensic pathology is critical in determining the cause and manner of death through post-mortem examinations, both macroscopic and microscopic. The field, however, grapples with issues such as outcome variability, laborious processes, and a scarcity of trained professionals. This paper presents SongCi, an innovative visual-language model (VLM) designed specifically for forensic pathology. SongCi utilizes advanced prototypical cross-modal self-supervised contrastive learning to enhance the accuracy, efficiency, and generalizability of forensic analyses. It was pre-trained and evaluated on a comprehensive multi-center dataset, which includes over 16 million high-resolution image patches, 2,228 vision-language pairs of post-mortem whole slide images (WSIs), and corresponding gross key findings, along with 471 distinct diagnostic outcomes. Our findings indicate that SongCi surpasses existing multi-modal AI models in many forensic pathology tasks, performs comparably to experienced forensic pathologists and significantly better than less experienced ones, and provides detailed multi-modal explainability, offering critical assistance in forensic investigations. To the best of our knowledge, SongCi is the first VLM specifically developed for forensic pathological analysis and the first large-vocabulary computational pathology (CPath) model that directly processes gigapixel WSIs in forensic science.
Punctate White Matter Lesion Segmentation in Preterm Infants Powered by Counterfactually Generative Learning
Sep 07, 2023Abstract:Accurate segmentation of punctate white matter lesions (PWMLs) are fundamental for the timely diagnosis and treatment of related developmental disorders. Automated PWMLs segmentation from infant brain MR images is challenging, considering that the lesions are typically small and low-contrast, and the number of lesions may dramatically change across subjects. Existing learning-based methods directly apply general network architectures to this challenging task, which may fail to capture detailed positional information of PWMLs, potentially leading to severe under-segmentations. In this paper, we propose to leverage the idea of counterfactual reasoning coupled with the auxiliary task of brain tissue segmentation to learn fine-grained positional and morphological representations of PWMLs for accurate localization and segmentation. A simple and easy-to-implement deep-learning framework (i.e., DeepPWML) is accordingly designed. It combines the lesion counterfactual map with the tissue probability map to train a lightweight PWML segmentation network, demonstrating state-of-the-art performance on a real-clinical dataset of infant T1w MR images. The code is available at \href{https://github.com/ladderlab-xjtu/DeepPWML}{https://github.com/ladderlab-xjtu/DeepPWML}.
Forensic Histopathological Recognition via a Context-Aware MIL Network Powered by Self-Supervised Contrastive Learning
Aug 27, 2023



Abstract:Forensic pathology is critical in analyzing death manner and time from the microscopic aspect to assist in the establishment of reliable factual bases for criminal investigation. In practice, even the manual differentiation between different postmortem organ tissues is challenging and relies on expertise, considering that changes like putrefaction and autolysis could significantly change typical histopathological appearance. Developing AI-based computational pathology techniques to assist forensic pathologists is practically meaningful, which requires reliable discriminative representation learning to capture tissues' fine-grained postmortem patterns. To this end, we propose a framework called FPath, in which a dedicated self-supervised contrastive learning strategy and a context-aware multiple-instance learning (MIL) block are designed to learn discriminative representations from postmortem histopathological images acquired at varying magnification scales. Our self-supervised learning step leverages multiple complementary contrastive losses and regularization terms to train a double-tier backbone for fine-grained and informative patch/instance embedding. Thereafter, the context-aware MIL adaptively distills from the local instances a holistic bag/image-level representation for the recognition task. On a large-scale database of $19,607$ experimental rat postmortem images and $3,378$ real-world human decedent images, our FPath led to state-of-the-art accuracy and promising cross-domain generalization in recognizing seven different postmortem tissues. The source code will be released on \href{https://github.com/ladderlab-xjtu/forensic_pathology}{https://github.com/ladderlab-xjtu/forensic\_pathology}.
Dual Meta-Learning with Longitudinally Generalized Regularization for One-Shot Brain Tissue Segmentation Across the Human Lifespan
Aug 13, 2023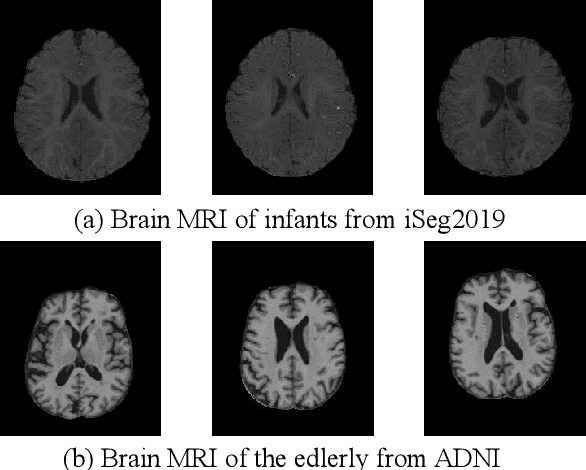

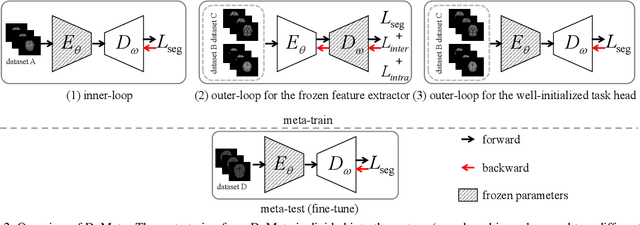
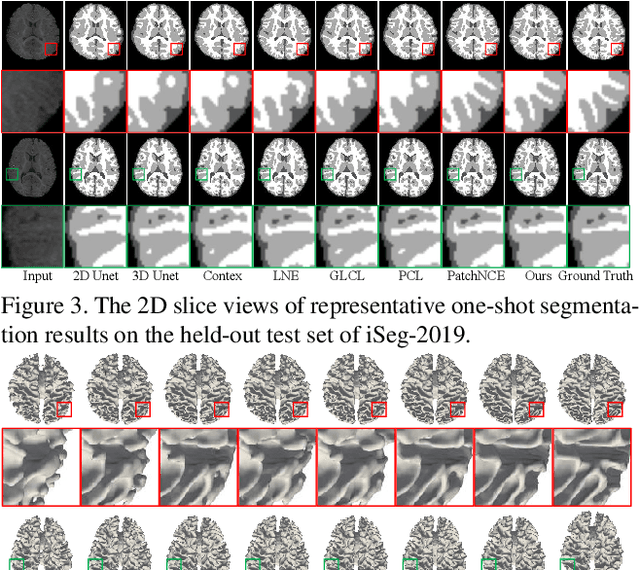
Abstract:Brain tissue segmentation is essential for neuroscience and clinical studies. However, segmentation on longitudinal data is challenging due to dynamic brain changes across the lifespan. Previous researches mainly focus on self-supervision with regularizations and will lose longitudinal generalization when fine-tuning on a specific age group. In this paper, we propose a dual meta-learning paradigm to learn longitudinally consistent representations and persist when fine-tuning. Specifically, we learn a plug-and-play feature extractor to extract longitudinal-consistent anatomical representations by meta-feature learning and a well-initialized task head for fine-tuning by meta-initialization learning. Besides, two class-aware regularizations are proposed to encourage longitudinal consistency. Experimental results on the iSeg2019 and ADNI datasets demonstrate the effectiveness of our method. Our code is available at https://github.com/ladderlab-xjtu/DuMeta.
NeuroExplainer: Fine-Grained Attention Decoding to Uncover Cortical Development Patterns of Preterm Infants
Jan 01, 2023Abstract:Deploying reliable deep learning techniques in interdisciplinary applications needs learned models to output accurate and ({even more importantly}) explainable predictions. Existing approaches typically explicate network outputs in a post-hoc fashion, under an implicit assumption that faithful explanations come from accurate predictions/classifications. We have an opposite claim that explanations boost (or even determine) classification. That is, end-to-end learning of explanation factors to augment discriminative representation extraction could be a more intuitive strategy to inversely assure fine-grained explainability, e.g., in those neuroimaging and neuroscience studies with high-dimensional data containing noisy, redundant, and task-irrelevant information. In this paper, we propose such an explainable geometric deep network dubbed as NeuroExplainer, with applications to uncover altered infant cortical development patterns associated with preterm birth. Given fundamental cortical attributes as network input, our NeuroExplainer adopts a hierarchical attention-decoding framework to learn fine-grained attentions and respective discriminative representations to accurately recognize preterm infants from term-born infants at term-equivalent age. NeuroExplainer learns the hierarchical attention-decoding modules under subject-level weak supervision coupled with targeted regularizers deduced from domain knowledge regarding brain development. These prior-guided constraints implicitly maximizes the explainability metrics (i.e., fidelity, sparsity, and stability) in network training, driving the learned network to output detailed explanations and accurate classifications. Experimental results on the public dHCP benchmark suggest that NeuroExplainer led to quantitatively reliable explanation results that are qualitatively consistent with representative neuroimaging studies.
Two-Stream Graph Convolutional Network for Intra-oral Scanner Image Segmentation
Apr 19, 2022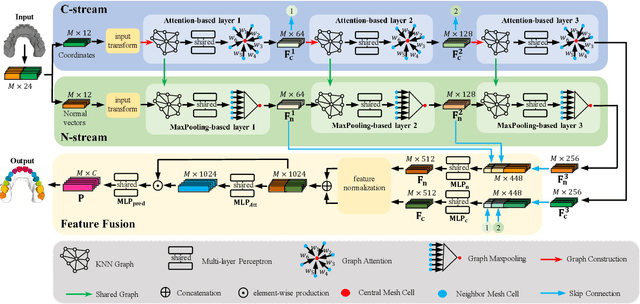
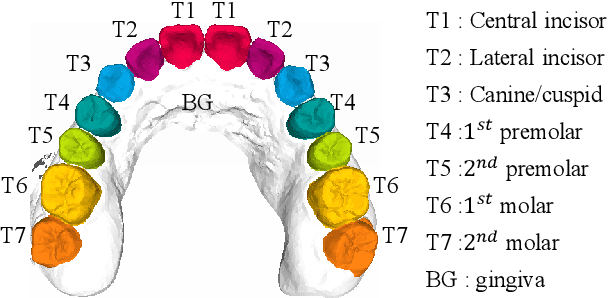
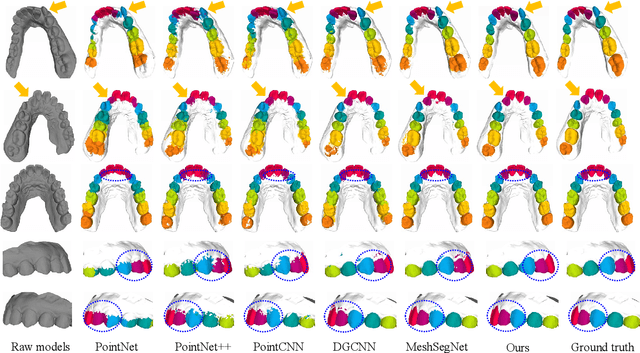
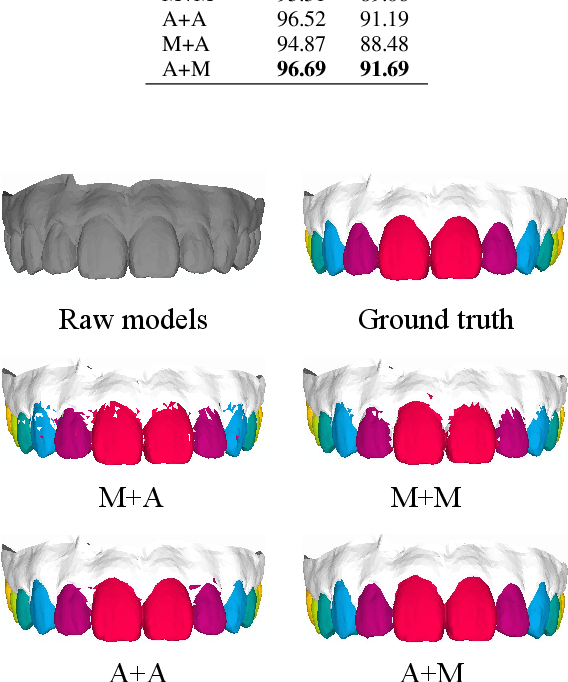
Abstract:Precise segmentation of teeth from intra-oral scanner images is an essential task in computer-aided orthodontic surgical planning. The state-of-the-art deep learning-based methods often simply concatenate the raw geometric attributes (i.e., coordinates and normal vectors) of mesh cells to train a single-stream network for automatic intra-oral scanner image segmentation. However, since different raw attributes reveal completely different geometric information, the naive concatenation of different raw attributes at the (low-level) input stage may bring unnecessary confusion in describing and differentiating between mesh cells, thus hampering the learning of high-level geometric representations for the segmentation task. To address this issue, we design a two-stream graph convolutional network (i.e., TSGCN), which can effectively handle inter-view confusion between different raw attributes to more effectively fuse their complementary information and learn discriminative multi-view geometric representations. Specifically, our TSGCN adopts two input-specific graph-learning streams to extract complementary high-level geometric representations from coordinates and normal vectors, respectively. Then, these single-view representations are further fused by a self-attention module to adaptively balance the contributions of different views in learning more discriminative multi-view representations for accurate and fully automatic tooth segmentation. We have evaluated our TSGCN on a real-patient dataset of dental (mesh) models acquired by 3D intraoral scanners. Experimental results show that our TSGCN significantly outperforms state-of-the-art methods in 3D tooth (surface) segmentation. Github: https://github.com/ZhangLingMing1/TSGCNet.
* 11 pages, 6 figures. arXiv admin note: text overlap with arXiv:2012.13697
SkullEngine: A Multi-stage CNN Framework for Collaborative CBCT Image Segmentation and Landmark Detection
Oct 07, 2021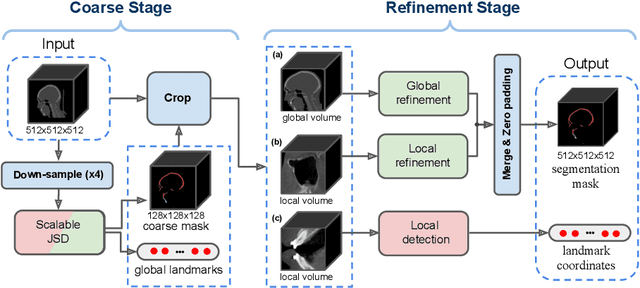
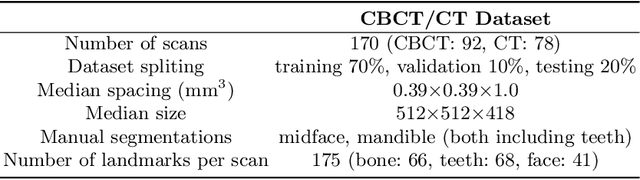
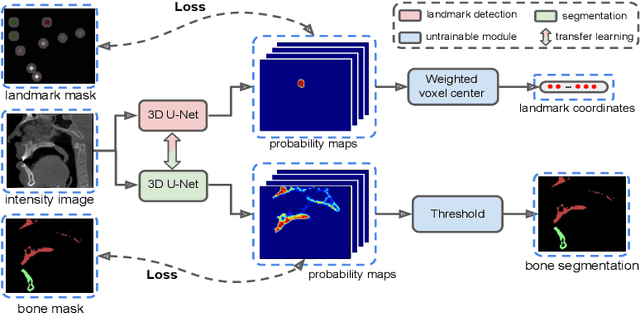
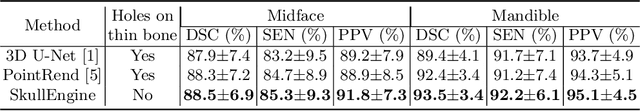
Abstract:We propose a multi-stage coarse-to-fine CNN-based framework, called SkullEngine, for high-resolution segmentation and large-scale landmark detection through a collaborative, integrated, and scalable JSD model and three segmentation and landmark detection refinement models. We evaluated our framework on a clinical dataset consisting of 170 CBCT/CT images for the task of segmenting 2 bones (midface and mandible) and detecting 175 clinically common landmarks on bones, teeth, and soft tissues.
Two-Stage Mesh Deep Learning for Automated Tooth Segmentation and Landmark Localization on 3D Intraoral Scans
Sep 24, 2021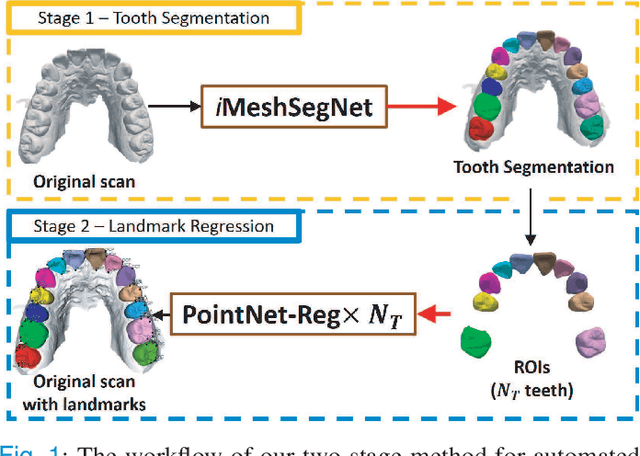
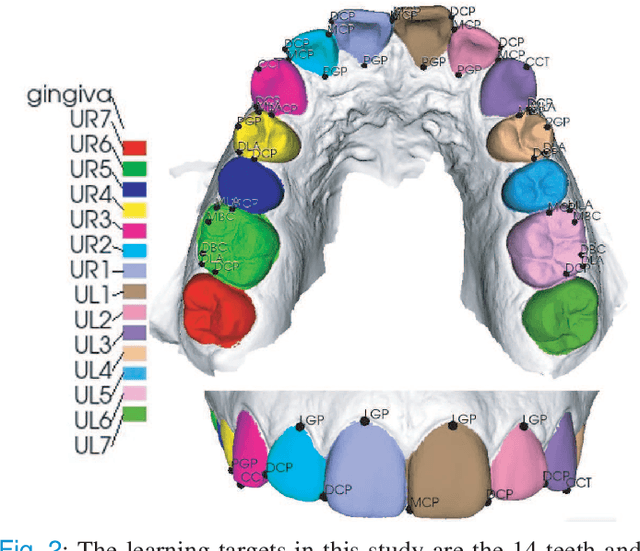
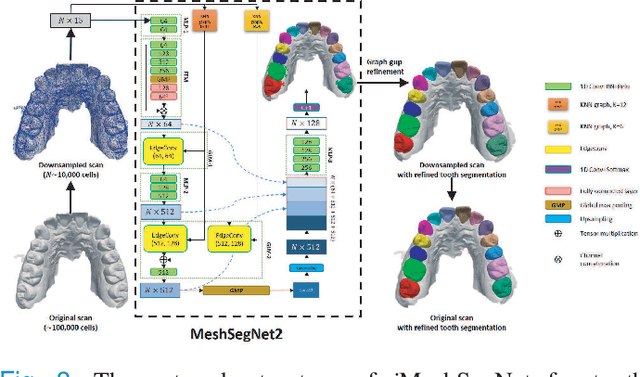
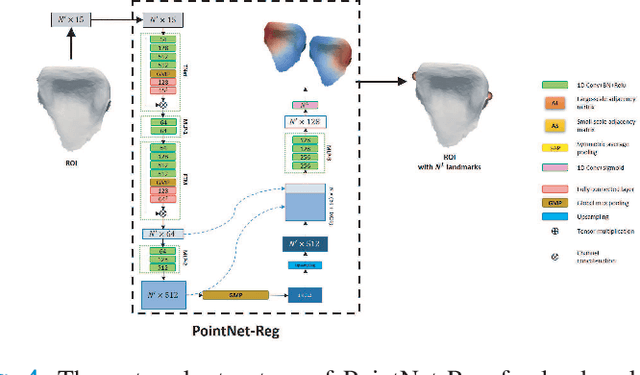
Abstract:Accurately segmenting teeth and identifying the corresponding anatomical landmarks on dental mesh models are essential in computer-aided orthodontic treatment. Manually performing these two tasks is time-consuming, tedious, and, more importantly, highly dependent on orthodontists' experiences due to the abnormality and large-scale variance of patients' teeth. Some machine learning-based methods have been designed and applied in the orthodontic field to automatically segment dental meshes (e.g., intraoral scans). In contrast, the number of studies on tooth landmark localization is still limited. This paper proposes a two-stage framework based on mesh deep learning (called TS-MDL) for joint tooth labeling and landmark identification on raw intraoral scans. Our TS-MDL first adopts an end-to-end \emph{i}MeshSegNet method (i.e., a variant of the existing MeshSegNet with both improved accuracy and efficiency) to label each tooth on the downsampled scan. Guided by the segmentation outputs, our TS-MDL further selects each tooth's region of interest (ROI) on the original mesh to construct a light-weight variant of the pioneering PointNet (i.e., PointNet-Reg) for regressing the corresponding landmark heatmaps. Our TS-MDL was evaluated on a real-clinical dataset, showing promising segmentation and localization performance. Specifically, \emph{i}MeshSegNet in the first stage of TS-MDL reached an averaged Dice similarity coefficient (DSC) at $0.953\pm0.076$, significantly outperforming the original MeshSegNet. In the second stage, PointNet-Reg achieved a mean absolute error (MAE) of $0.623\pm0.718 \, mm$ in distances between the prediction and ground truth for $44$ landmarks, which is superior compared with other networks for landmark detection. All these results suggest the potential usage of our TS-MDL in clinical practices.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge