Hongtu Zhu
Parallel-Probe: Towards Efficient Parallel Thinking via 2D Probing
Feb 03, 2026Abstract:Parallel thinking has emerged as a promising paradigm for reasoning, yet it imposes significant computational burdens. Existing efficiency methods primarily rely on local, per-trajectory signals and lack principled mechanisms to exploit global dynamics across parallel branches. We introduce 2D probing, an interface that exposes the width-depth dynamics of parallel thinking by periodically eliciting intermediate answers from all branches. Our analysis reveals three key insights: non-monotonic scaling across width-depth allocations, heterogeneous reasoning branch lengths, and early stabilization of global consensus. Guided by these insights, we introduce $\textbf{Parallel-Probe}$, a training-free controller designed to optimize online parallel thinking. Parallel-Probe employs consensus-based early stopping to regulate reasoning depth and deviation-based branch pruning to dynamically adjust width. Extensive experiments across three benchmarks and multiple models demonstrate that Parallel-Probe establishes a superior Pareto frontier for test-time scaling. Compared to standard majority voting, it reduces sequential tokens by up to $\textbf{35.8}$% and total token cost by over $\textbf{25.8}$% while maintaining competitive accuracy.
Designing Time Series Experiments in A/B Testing with Transformer Reinforcement Learning
Feb 02, 2026Abstract:A/B testing has become a gold standard for modern technological companies to conduct policy evaluation. Yet, its application to time series experiments, where policies are sequentially assigned over time, remains challenging. Existing designs suffer from two limitations: (i) they do not fully leverage the entire history for treatment allocation; (ii) they rely on strong assumptions to approximate the objective function (e.g., the mean squared error of the estimated treatment effect) for optimizing the design. We first establish an impossibility theorem showing that failure to condition on the full history leads to suboptimal designs, due to the dynamic dependencies in time series experiments. To address both limitations simultaneously, we next propose a transformer reinforcement learning (RL) approach which leverages transformers to condition allocation on the entire history and employs RL to directly optimize the MSE without relying on restrictive assumptions. Empirical evaluations on synthetic data, a publicly available dispatch simulator, and a real-world ridesharing dataset demonstrate that our proposal consistently outperforms existing designs.
"Rebuilding" Statistics in the Age of AI: A Town Hall Discussion on Culture, Infrastructure, and Training
Jan 24, 2026Abstract:This article presents the full, original record of the 2024 Joint Statistical Meetings (JSM) town hall, "Statistics in the Age of AI," which convened leading statisticians to discuss how the field is evolving in response to advances in artificial intelligence, foundation models, large-scale empirical modeling, and data-intensive infrastructures. The town hall was structured around open panel discussion and extensive audience Q&A, with the aim of eliciting candid, experience-driven perspectives rather than formal presentations or prepared statements. This document preserves the extended exchanges among panelists and audience members, with minimal editorial intervention, and organizes the conversation around five recurring questions concerning disciplinary culture and practices, data curation and "data work," engagement with modern empirical modeling, training for large-scale AI applications, and partnerships with key AI stakeholders. By providing an archival record of this discussion, the preprint aims to support transparency, community reflection, and ongoing dialogue about the evolving role of statistics in the data- and AI-centric future.
CDE: Curiosity-Driven Exploration for Efficient Reinforcement Learning in Large Language Models
Sep 11, 2025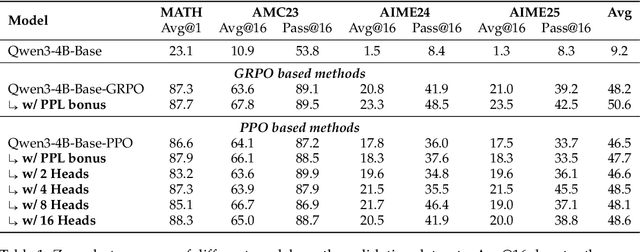
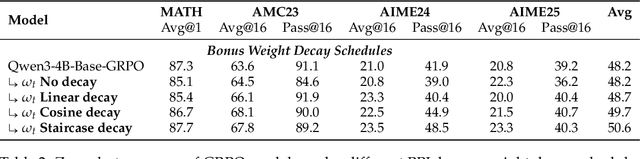
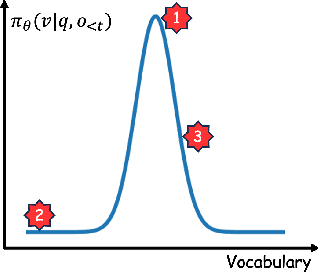
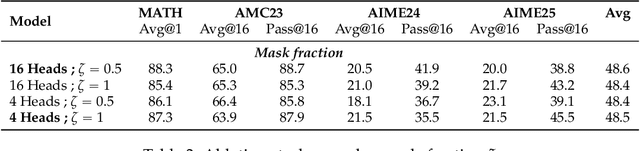
Abstract:Reinforcement Learning with Verifiable Rewards (RLVR) is a powerful paradigm for enhancing the reasoning ability of Large Language Models (LLMs). Yet current RLVR methods often explore poorly, leading to premature convergence and entropy collapse. To address this challenge, we introduce Curiosity-Driven Exploration (CDE), a framework that leverages the model's own intrinsic sense of curiosity to guide exploration. We formalize curiosity with signals from both the actor and the critic: for the actor, we use perplexity over its generated response, and for the critic, we use the variance of value estimates from a multi-head architecture. Both signals serve as an exploration bonus within the RLVR framework to guide the model. Our theoretical analysis shows that the actor-wise bonus inherently penalizes overconfident errors and promotes diversity among correct responses; moreover, we connect the critic-wise bonus to the well-established count-based exploration bonus in RL. Empirically, our method achieves an approximate +3 point improvement over standard RLVR using GRPO/PPO on AIME benchmarks. Further analysis identifies a calibration collapse mechanism within RLVR, shedding light on common LLM failure modes.
High-Dimensional Dynamic Covariance Models with Random Forests
May 18, 2025Abstract:This paper introduces a novel nonparametric method for estimating high-dimensional dynamic covariance matrices with multiple conditioning covariates, leveraging random forests and supported by robust theoretical guarantees. Unlike traditional static methods, our dynamic nonparametric covariance models effectively capture distributional heterogeneity. Furthermore, unlike kernel-smoothing methods, which are restricted to a single conditioning covariate, our approach accommodates multiple covariates in a fully nonparametric framework. To the best of our knowledge, this is the first method to use random forests for estimating high-dimensional dynamic covariance matrices. In high-dimensional settings, we establish uniform consistency theory, providing nonasymptotic error rates and model selection properties, even when the response dimension grows sub-exponentially with the sample size. These results hold uniformly across a range of conditioning variables. The method's effectiveness is demonstrated through simulations and a stock dataset analysis, highlighting its ability to model complex dynamics in high-dimensional scenarios.
Deep Distributional Learning with Non-crossing Quantile Network
Apr 11, 2025Abstract:In this paper, we introduce a non-crossing quantile (NQ) network for conditional distribution learning. By leveraging non-negative activation functions, the NQ network ensures that the learned distributions remain monotonic, effectively addressing the issue of quantile crossing. Furthermore, the NQ network-based deep distributional learning framework is highly adaptable, applicable to a wide range of applications, from classical non-parametric quantile regression to more advanced tasks such as causal effect estimation and distributional reinforcement learning (RL). We also develop a comprehensive theoretical foundation for the deep NQ estimator and its application to distributional RL, providing an in-depth analysis that demonstrates its effectiveness across these domains. Our experimental results further highlight the robustness and versatility of the NQ network.
Spatio-temporal Prediction of Fine-Grained Origin-Destination Matrices with Applications in Ridesharing
Mar 31, 2025Abstract:Accurate spatial-temporal prediction of network-based travelers' requests is crucial for the effective policy design of ridesharing platforms. Having knowledge of the total demand between various locations in the upcoming time slots enables platforms to proactively prepare adequate supplies, thereby increasing the likelihood of fulfilling travelers' requests and redistributing idle drivers to areas with high potential demand to optimize the global supply-demand equilibrium. This paper delves into the prediction of Origin-Destination (OD) demands at a fine-grained spatial level, especially when confronted with an expansive set of local regions. While this task holds immense practical value, it remains relatively unexplored within the research community. To fill this gap, we introduce a novel prediction model called OD-CED, which comprises an unsupervised space coarsening technique to alleviate data sparsity and an encoder-decoder architecture to capture both semantic and geographic dependencies. Through practical experimentation, OD-CED has demonstrated remarkable results. It achieved an impressive reduction of up to 45% reduction in root-mean-square error and 60% in weighted mean absolute percentage error over traditional statistical methods when dealing with OD matrices exhibiting a sparsity exceeding 90%.
MDocAgent: A Multi-Modal Multi-Agent Framework for Document Understanding
Mar 18, 2025Abstract:Document Question Answering (DocQA) is a very common task. Existing methods using Large Language Models (LLMs) or Large Vision Language Models (LVLMs) and Retrieval Augmented Generation (RAG) often prioritize information from a single modal, failing to effectively integrate textual and visual cues. These approaches struggle with complex multi-modal reasoning, limiting their performance on real-world documents. We present MDocAgent (A Multi-Modal Multi-Agent Framework for Document Understanding), a novel RAG and multi-agent framework that leverages both text and image. Our system employs five specialized agents: a general agent, a critical agent, a text agent, an image agent and a summarizing agent. These agents engage in multi-modal context retrieval, combining their individual insights to achieve a more comprehensive understanding of the document's content. This collaborative approach enables the system to synthesize information from both textual and visual components, leading to improved accuracy in question answering. Preliminary experiments on five benchmarks like MMLongBench, LongDocURL demonstrate the effectiveness of our MDocAgent, achieve an average improvement of 12.1% compared to current state-of-the-art method. This work contributes to the development of more robust and comprehensive DocQA systems capable of handling the complexities of real-world documents containing rich textual and visual information. Our data and code are available at https://github.com/aiming-lab/MDocAgent.
Large Language Models for Bioinformatics
Jan 10, 2025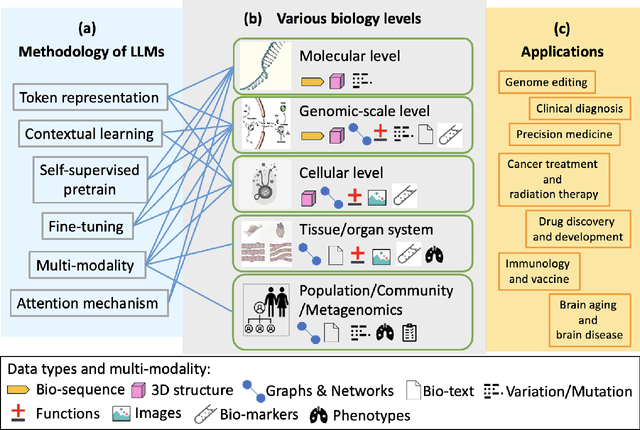
Abstract:With the rapid advancements in large language model (LLM) technology and the emergence of bioinformatics-specific language models (BioLMs), there is a growing need for a comprehensive analysis of the current landscape, computational characteristics, and diverse applications. This survey aims to address this need by providing a thorough review of BioLMs, focusing on their evolution, classification, and distinguishing features, alongside a detailed examination of training methodologies, datasets, and evaluation frameworks. We explore the wide-ranging applications of BioLMs in critical areas such as disease diagnosis, drug discovery, and vaccine development, highlighting their impact and transformative potential in bioinformatics. We identify key challenges and limitations inherent in BioLMs, including data privacy and security concerns, interpretability issues, biases in training data and model outputs, and domain adaptation complexities. Finally, we highlight emerging trends and future directions, offering valuable insights to guide researchers and clinicians toward advancing BioLMs for increasingly sophisticated biological and clinical applications.
MMedPO: Aligning Medical Vision-Language Models with Clinical-Aware Multimodal Preference Optimization
Dec 09, 2024



Abstract:The advancement of Large Vision-Language Models (LVLMs) has propelled their application in the medical field. However, Medical LVLMs (Med-LVLMs) encounter factuality challenges due to modality misalignment, where the models prioritize textual knowledge over visual input, leading to hallucinations that contradict information in medical images. Previous attempts to enhance modality alignment in Med-LVLMs through preference optimization have inadequately mitigated clinical relevance in preference data, making these samples easily distinguishable and reducing alignment effectiveness. To address this challenge, we propose MMedPO, a novel multimodal medical preference optimization approach that considers the clinical relevance of preference samples to enhance Med-LVLM alignment. MMedPO curates multimodal preference data by introducing two types of dispreference: (1) plausible hallucinations injected through target Med-LVLMs or GPT-4o to produce medically inaccurate responses, and (2) lesion region neglect achieved through local lesion-noising, disrupting visual understanding of critical areas. We then calculate clinical relevance for each sample based on scores from multiple Med-LLMs and visual tools, and integrate these scores into the preference optimization process as weights, enabling effective alignment. Our experiments demonstrate that MMedPO significantly enhances factual accuracy in Med-LVLMs, achieving substantial improvements over existing preference optimization methods by averaging 14.2% and 51.7% across the Med-VQA and report generation tasks. Our code are available in https://github.com/aiming-lab/MMedPO.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge