Jiameng Liu
Towards General Text-guided Image Synthesis for Customized Multimodal Brain MRI Generation
Sep 25, 2024



Abstract:Multimodal brain magnetic resonance (MR) imaging is indispensable in neuroscience and neurology. However, due to the accessibility of MRI scanners and their lengthy acquisition time, multimodal MR images are not commonly available. Current MR image synthesis approaches are typically trained on independent datasets for specific tasks, leading to suboptimal performance when applied to novel datasets and tasks. Here, we present TUMSyn, a Text-guided Universal MR image Synthesis generalist model, which can flexibly generate brain MR images with demanded imaging metadata from routinely acquired scans guided by text prompts. To ensure TUMSyn's image synthesis precision, versatility, and generalizability, we first construct a brain MR database comprising 31,407 3D images with 7 MRI modalities from 13 centers. We then pre-train an MRI-specific text encoder using contrastive learning to effectively control MR image synthesis based on text prompts. Extensive experiments on diverse datasets and physician assessments indicate that TUMSyn can generate clinically meaningful MR images with specified imaging metadata in supervised and zero-shot scenarios. Therefore, TUMSyn can be utilized along with acquired MR scan(s) to facilitate large-scale MRI-based screening and diagnosis of brain diseases.
SDF-Net: A Hybrid Detection Network for Mediastinal Lymph Node Detection on Contrast CT Images
Sep 10, 2024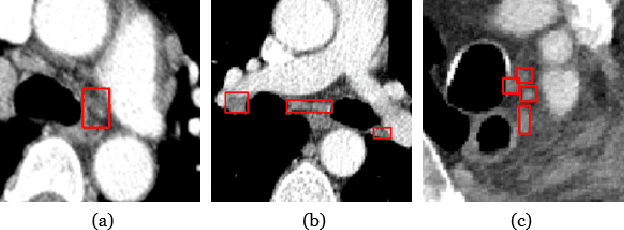
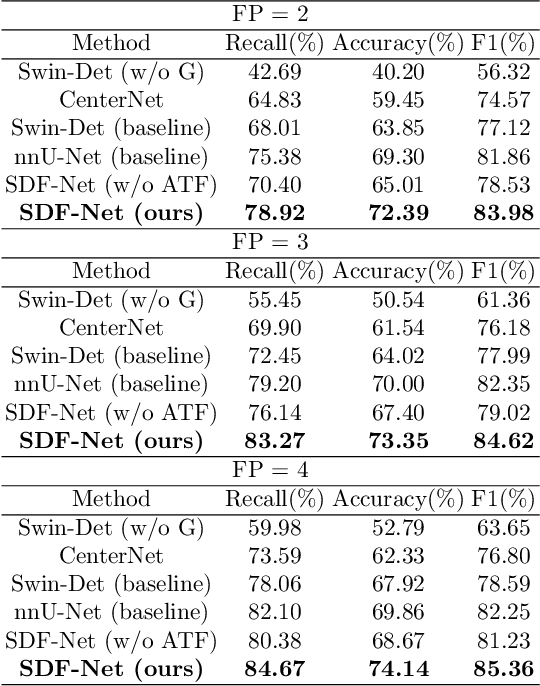
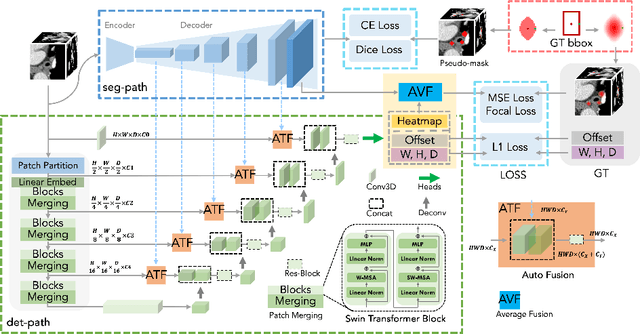
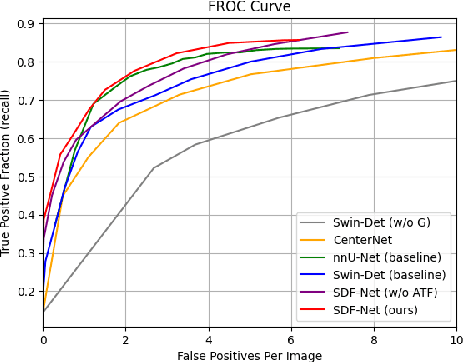
Abstract:Accurate lymph node detection and quantification are crucial for cancer diagnosis and staging on contrast-enhanced CT images, as they impact treatment planning and prognosis. However, detecting lymph nodes in the mediastinal area poses challenges due to their low contrast, irregular shapes and dispersed distribution. In this paper, we propose a Swin-Det Fusion Network (SDF-Net) to effectively detect lymph nodes. SDF-Net integrates features from both segmentation and detection to enhance the detection capability of lymph nodes with various shapes and sizes. Specifically, an auto-fusion module is designed to merge the feature maps of segmentation and detection networks at different levels. To facilitate effective learning without mask annotations, we introduce a shape-adaptive Gaussian kernel to represent lymph node in the training stage and provide more anatomical information for effective learning. Comparative results demonstrate promising performance in addressing the complex lymph node detection problem.
Continuous Invariance Learning
Oct 09, 2023



Abstract:Invariance learning methods aim to learn invariant features in the hope that they generalize under distributional shifts. Although many tasks are naturally characterized by continuous domains, current invariance learning techniques generally assume categorically indexed domains. For example, auto-scaling in cloud computing often needs a CPU utilization prediction model that generalizes across different times (e.g., time of a day and date of a year), where `time' is a continuous domain index. In this paper, we start by theoretically showing that existing invariance learning methods can fail for continuous domain problems. Specifically, the naive solution of splitting continuous domains into discrete ones ignores the underlying relationship among domains, and therefore potentially leads to suboptimal performance. To address this challenge, we then propose Continuous Invariance Learning (CIL), which extracts invariant features across continuously indexed domains. CIL is a novel adversarial procedure that measures and controls the conditional independence between the labels and continuous domain indices given the extracted features. Our theoretical analysis demonstrates the superiority of CIL over existing invariance learning methods. Empirical results on both synthetic and real-world datasets (including data collected from production systems) show that CIL consistently outperforms strong baselines among all the tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge