Jiliu Zhou
Large Language Models for Single-Step and Multi-Step Flight Trajectory Prediction
Jan 29, 2025Abstract:Flight trajectory prediction is a critical time series task in aviation. While deep learning methods have shown significant promise, the application of large language models (LLMs) to this domain remains underexplored. This study pioneers the use of LLMs for flight trajectory prediction by reframing it as a language modeling problem. Specifically, We extract features representing the aircraft's position and status from ADS-B flight data to construct a prompt-based dataset, where trajectory waypoints are converted into language tokens. The dataset is then employed to fine-tune LLMs, enabling them to learn complex spatiotemporal patterns for accurate predictions. Comprehensive experiments demonstrate that LLMs achieve notable performance improvements in both single-step and multi-step predictions compared to traditional methods, with LLaMA-3.1 model achieving the highest overall accuracy. However, the high inference latency of LLMs poses a challenge for real-time applications, underscoring the need for further research in this promising direction.
BASIC: Semi-supervised Multi-organ Segmentation with Balanced Subclass Regularization and Semantic-conflict Penalty
Jan 07, 2025



Abstract:Semi-supervised learning (SSL) has shown notable potential in relieving the heavy demand of dense prediction tasks on large-scale well-annotated datasets, especially for the challenging multi-organ segmentation (MoS). However, the prevailing class-imbalance problem in MoS caused by the substantial variations in organ size exacerbates the learning difficulty of the SSL network. To address this issue, in this paper, we propose an innovative semi-supervised network with BAlanced Subclass regularIzation and semantic-Conflict penalty mechanism (BASIC) to effectively learn the unbiased knowledge for semi-supervised MoS. Concretely, we construct a novel auxiliary subclass segmentation (SCS) task based on priorly generated balanced subclasses, thus deeply excavating the unbiased information for the main MoS task with the fashion of multi-task learning. Additionally, based on a mean teacher framework, we elaborately design a balanced subclass regularization to utilize the teacher predictions of SCS task to supervise the student predictions of MoS task, thus effectively transferring unbiased knowledge to the MoS subnetwork and alleviating the influence of the class-imbalance problem. Considering the similar semantic information inside the subclasses and their corresponding original classes (i.e., parent classes), we devise a semantic-conflict penalty mechanism to give heavier punishments to the conflicting SCS predictions with wrong parent classes and provide a more accurate constraint to the MoS predictions. Extensive experiments conducted on two publicly available datasets, i.e., the WORD dataset and the MICCAI FLARE 2022 dataset, have verified the superior performance of our proposed BASIC compared to other state-of-the-art methods.
BTMuda: A Bi-level Multi-source unsupervised domain adaptation framework for breast cancer diagnosis
Aug 30, 2024Abstract:Deep learning has revolutionized the early detection of breast cancer, resulting in a significant decrease in mortality rates. However, difficulties in obtaining annotations and huge variations in distribution between training sets and real scenes have limited their clinical applications. To address these limitations, unsupervised domain adaptation (UDA) methods have been used to transfer knowledge from one labeled source domain to the unlabeled target domain, yet these approaches suffer from severe domain shift issues and often ignore the potential benefits of leveraging multiple relevant sources in practical applications. To address these limitations, in this work, we construct a Three-Branch Mixed extractor and propose a Bi-level Multi-source unsupervised domain adaptation method called BTMuda for breast cancer diagnosis. Our method addresses the problems of domain shift by dividing domain shift issues into two levels: intra-domain and inter-domain. To reduce the intra-domain shift, we jointly train a CNN and a Transformer as two paths of a domain mixed feature extractor to obtain robust representations rich in both low-level local and high-level global information. As for the inter-domain shift, we redesign the Transformer delicately to a three-branch architecture with cross-attention and distillation, which learns domain-invariant representations from multiple domains. Besides, we introduce two alignment modules - one for feature alignment and one for classifier alignment - to improve the alignment process. Extensive experiments conducted on three public mammographic datasets demonstrate that our BTMuda outperforms state-of-the-art methods.
S3PET: Semi-supervised Standard-dose PET Image Reconstruction via Dose-aware Token Swap
Jul 30, 2024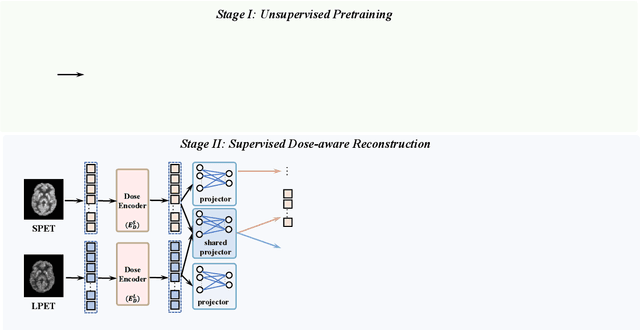
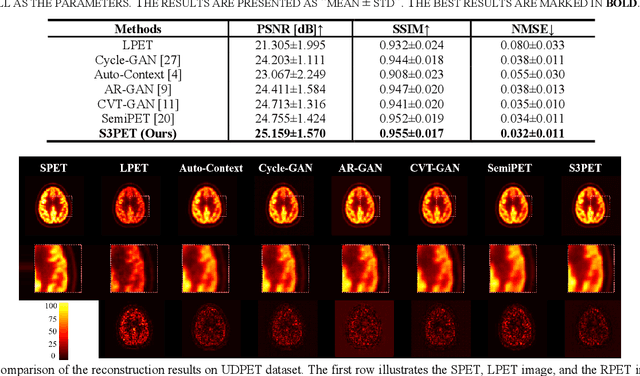
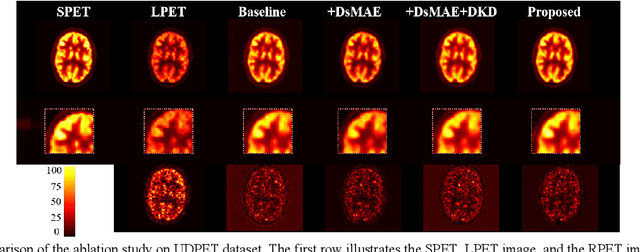
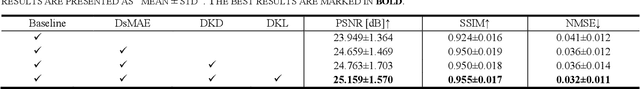
Abstract:To acquire high-quality positron emission tomography (PET) images while reducing the radiation tracer dose, numerous efforts have been devoted to reconstructing standard-dose PET (SPET) images from low-dose PET (LPET). However, the success of current fully-supervised approaches relies on abundant paired LPET and SPET images, which are often unavailable in clinic. Moreover, these methods often mix the dose-invariant content with dose level-related dose-specific details during reconstruction, resulting in distorted images. To alleviate these problems, in this paper, we propose a two-stage Semi-Supervised SPET reconstruction framework, namely S3PET, to accommodate the training of abundant unpaired and limited paired SPET and LPET images. Our S3PET involves an un-supervised pre-training stage (Stage I) to extract representations from unpaired images, and a supervised dose-aware reconstruction stage (Stage II) to achieve LPET-to-SPET reconstruction by transferring the dose-specific knowledge between paired images. Specifically, in stage I, two independent dose-specific masked autoencoders (DsMAEs) are adopted to comprehensively understand the unpaired SPET and LPET images. Then, in Stage II, the pre-trained DsMAEs are further finetuned using paired images. To prevent distortions in both content and details, we introduce two elaborate modules, i.e., a dose knowledge decouple module to disentangle the respective dose-specific and dose-invariant knowledge of LPET and SPET, and a dose-specific knowledge learning module to transfer the dose-specific information from SPET to LPET, thereby achieving high-quality SPET reconstruction from LPET images. Experiments on two datasets demonstrate that our S3PET achieves state-of-the-art performance quantitatively and qualitatively.
Learning with Alignments: Tackling the Inter- and Intra-domain Shifts for Cross-multidomain Facial Expression Recognition
Jul 08, 2024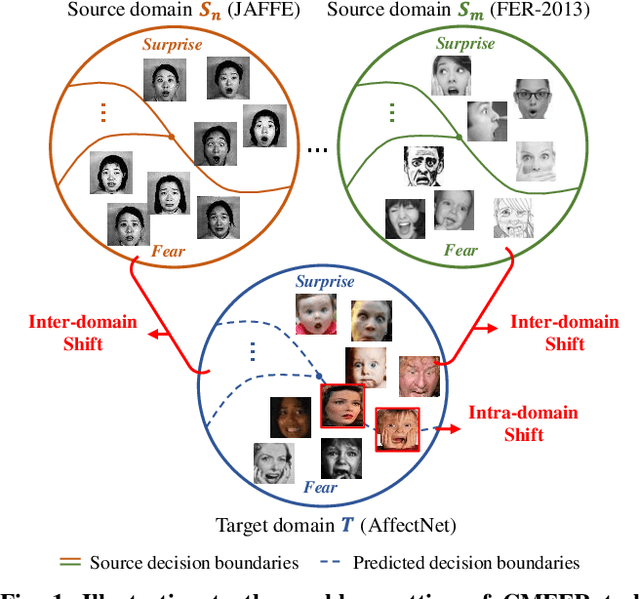

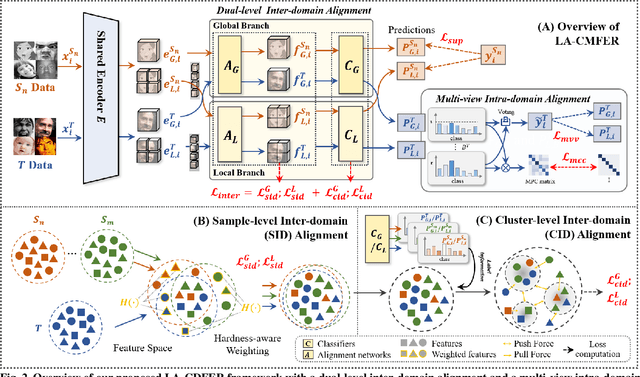
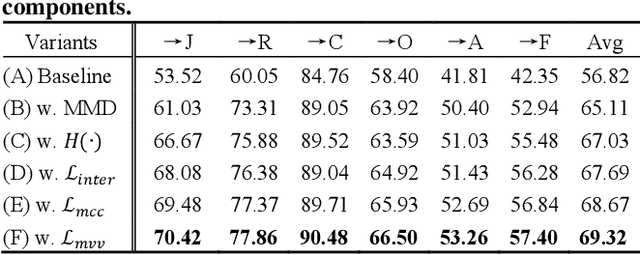
Abstract:Facial Expression Recognition (FER) holds significant importance in human-computer interactions. Existing cross-domain FER methods often transfer knowledge solely from a single labeled source domain to an unlabeled target domain, neglecting the comprehensive information across multiple sources. Nevertheless, cross-multidomain FER (CMFER) is very challenging for (i) the inherent inter-domain shifts across multiple domains and (ii) the intra-domain shifts stemming from the ambiguous expressions and low inter-class distinctions. In this paper, we propose a novel Learning with Alignments CMFER framework, named LA-CMFER, to handle both inter- and intra-domain shifts. Specifically, LA-CMFER is constructed with a global branch and a local branch to extract features from the full images and local subtle expressions, respectively. Based on this, LA-CMFER presents a dual-level inter-domain alignment method to force the model to prioritize hard-to-align samples in knowledge transfer at a sample level while gradually generating a well-clustered feature space with the guidance of class attributes at a cluster level, thus narrowing the inter-domain shifts. To address the intra-domain shifts, LA-CMFER introduces a multi-view intra-domain alignment method with a multi-view clustering consistency constraint where a prediction similarity matrix is built to pursue consistency between the global and local views, thus refining pseudo labels and eliminating latent noise. Extensive experiments on six benchmark datasets have validated the superiority of our LA-CMFER.
MCAD: Multi-modal Conditioned Adversarial Diffusion Model for High-Quality PET Image Reconstruction
Jun 19, 2024Abstract:Radiation hazards associated with standard-dose positron emission tomography (SPET) images remain a concern, whereas the quality of low-dose PET (LPET) images fails to meet clinical requirements. Therefore, there is great interest in reconstructing SPET images from LPET images. However, prior studies focus solely on image data, neglecting vital complementary information from other modalities, e.g., patients' clinical tabular, resulting in compromised reconstruction with limited diagnostic utility. Moreover, they often overlook the semantic consistency between real SPET and reconstructed images, leading to distorted semantic contexts. To tackle these problems, we propose a novel Multi-modal Conditioned Adversarial Diffusion model (MCAD) to reconstruct SPET images from multi-modal inputs, including LPET images and clinical tabular. Specifically, our MCAD incorporates a Multi-modal conditional Encoder (Mc-Encoder) to extract multi-modal features, followed by a conditional diffusion process to blend noise with multi-modal features and gradually map blended features to the target SPET images. To balance multi-modal inputs, the Mc-Encoder embeds Optimal Multi-modal Transport co-Attention (OMTA) to narrow the heterogeneity gap between image and tabular while capturing their interactions, providing sufficient guidance for reconstruction. In addition, to mitigate semantic distortions, we introduce the Multi-Modal Masked Text Reconstruction (M3TRec), which leverages semantic knowledge extracted from denoised PET images to restore the masked clinical tabular, thereby compelling the network to maintain accurate semantics during reconstruction. To expedite the diffusion process, we further introduce an adversarial diffusive network with a reduced number of diffusion steps. Experiments show that our method achieves the state-of-the-art performance both qualitatively and quantitatively.
Adaptive Prompt Learning with Negative Textual Semantics and Uncertainty Modeling for Universal Multi-Source Domain Adaptation
Apr 24, 2024Abstract:Universal Multi-source Domain Adaptation (UniMDA) transfers knowledge from multiple labeled source domains to an unlabeled target domain under domain shifts (different data distribution) and class shifts (unknown target classes). Existing solutions focus on excavating image features to detect unknown samples, ignoring abundant information contained in textual semantics. In this paper, we propose an Adaptive Prompt learning with Negative textual semantics and uncErtainty modeling method based on Contrastive Language-Image Pre-training (APNE-CLIP) for UniMDA classification tasks. Concretely, we utilize the CLIP with adaptive prompts to leverage textual information of class semantics and domain representations, helping the model identify unknown samples and address domain shifts. Additionally, we design a novel global instance-level alignment objective by utilizing negative textual semantics to achieve more precise image-text pair alignment. Furthermore, we propose an energy-based uncertainty modeling strategy to enlarge the margin distance between known and unknown samples. Extensive experiments demonstrate the superiority of our proposed method.
Two-Phase Multi-Dose-Level PET Image Reconstruction with Dose Level Awareness
Apr 10, 2024Abstract:To obtain high-quality positron emission tomography (PET) while minimizing radiation exposure, a range of methods have been designed to reconstruct standard-dose PET (SPET) from corresponding low-dose PET (LPET) images. However, most current methods merely learn the mapping between single-dose-level LPET and SPET images, but omit the dose disparity of LPET images in clinical scenarios. In this paper, to reconstruct high-quality SPET images from multi-dose-level LPET images, we design a novel two-phase multi-dose-level PET reconstruction algorithm with dose level awareness, containing a pre-training phase and a SPET prediction phase. Specifically, the pre-training phase is devised to explore both fine-grained discriminative features and effective semantic representation. The SPET prediction phase adopts a coarse prediction network utilizing pre-learned dose level prior to generate preliminary result, and a refinement network to precisely preserve the details. Experiments on MICCAI 2022 Ultra-low Dose PET Imaging Challenge Dataset have demonstrated the superiority of our method.
Dcl-Net: Dual Contrastive Learning Network for Semi-Supervised Multi-Organ Segmentation
Mar 06, 2024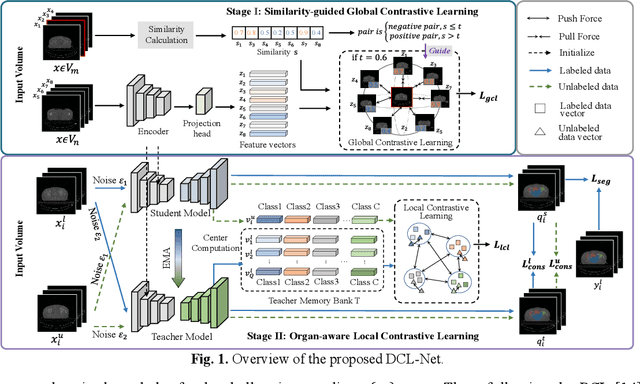

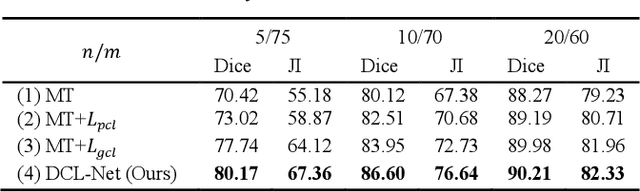
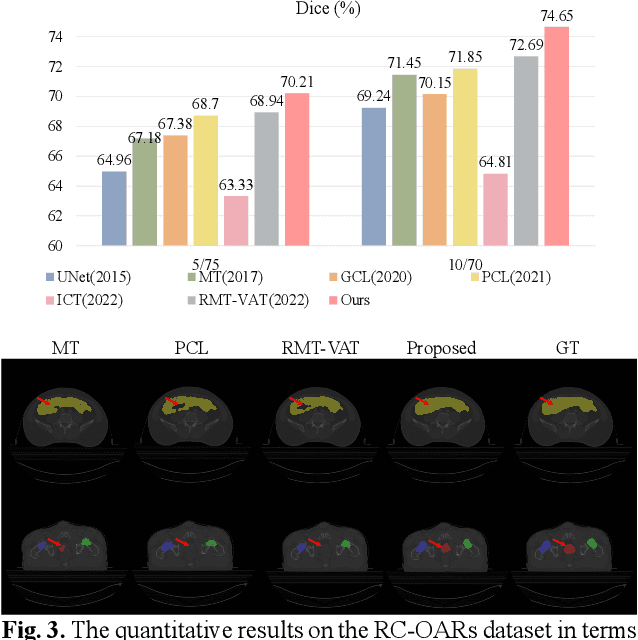
Abstract:Semi-supervised learning is a sound measure to relieve the strict demand of abundant annotated datasets, especially for challenging multi-organ segmentation . However, most existing SSL methods predict pixels in a single image independently, ignoring the relations among images and categories. In this paper, we propose a two-stage Dual Contrastive Learning Network for semi-supervised MoS, which utilizes global and local contrastive learning to strengthen the relations among images and classes. Concretely, in Stage 1, we develop a similarity-guided global contrastive learning to explore the implicit continuity and similarity among images and learn global context. Then, in Stage 2, we present an organ-aware local contrastive learning to further attract the class representations. To ease the computation burden, we introduce a mask center computation algorithm to compress the category representations for local contrastive learning. Experiments conducted on the public 2017 ACDC dataset and an in-house RC-OARs dataset has demonstrated the superior performance of our method.
Dose Prediction Driven Radiotherapy Paramters Regression via Intra- and Inter-Relation Modeling
Feb 29, 2024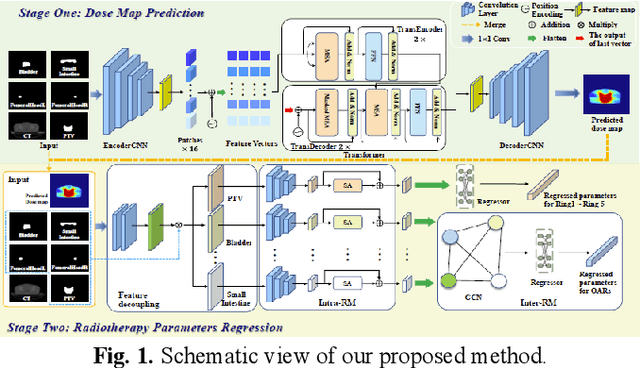
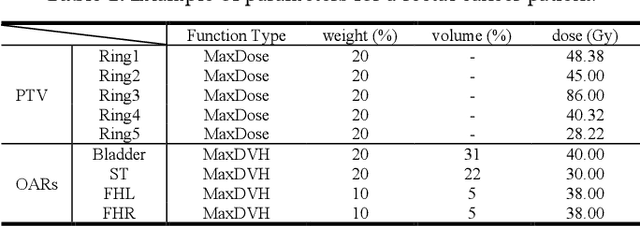
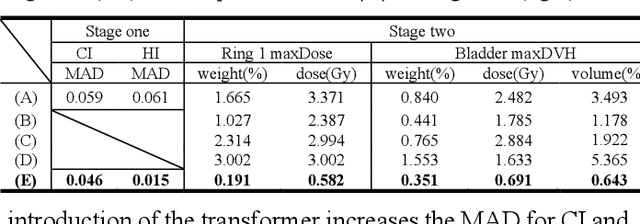

Abstract:Deep learning has facilitated the automation of radiotherapy by predicting accurate dose distribution maps. However, existing methods fail to derive the desirable radiotherapy parameters that can be directly input into the treatment planning system (TPS), impeding the full automation of radiotherapy. To enable more thorough automatic radiotherapy, in this paper, we propose a novel two-stage framework to directly regress the radiotherapy parameters, including a dose map prediction stage and a radiotherapy parameters regression stage. In stage one, we combine transformer and convolutional neural network (CNN) to predict realistic dose maps with rich global and local information, providing accurate dosimetric knowledge for the subsequent parameters regression. In stage two, two elaborate modules, i.e., an intra-relation modeling (Intra-RM) module and an inter-relation modeling (Inter-RM) module, are designed to exploit the organ-specific and organ-shared features for precise parameters regression. Experimental results on a rectal cancer dataset demonstrate the effectiveness of our method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge