Binyu Yan
BAR: A Backward Reasoning based Agent for Complex Minecraft Tasks
May 20, 2025



Abstract:Large language model (LLM) based agents have shown great potential in following human instructions and automatically completing various tasks. To complete a task, the agent needs to decompose it into easily executed steps by planning. Existing studies mainly conduct the planning by inferring what steps should be executed next starting from the agent's initial state. However, this forward reasoning paradigm doesn't work well for complex tasks. We propose to study this issue in Minecraft, a virtual environment that simulates complex tasks based on real-world scenarios. We believe that the failure of forward reasoning is caused by the big perception gap between the agent's initial state and task goal. To this end, we leverage backward reasoning and make the planning starting from the terminal state, which can directly achieve the task goal in one step. Specifically, we design a BAckward Reasoning based agent (BAR). It is equipped with a recursive goal decomposition module, a state consistency maintaining module and a stage memory module to make robust, consistent, and efficient planning starting from the terminal state. Experimental results demonstrate the superiority of BAR over existing methods and the effectiveness of proposed modules.
MIND: Towards Immersive Psychological Healing with Multi-agent Inner Dialogue
Feb 27, 2025Abstract:Mental health issues are worsening in today's competitive society, such as depression and anxiety. Traditional healings like counseling and chatbots fail to engage effectively, they often provide generic responses lacking emotional depth. Although large language models (LLMs) have the potential to create more human-like interactions, they still struggle to capture subtle emotions. This requires LLMs to be equipped with human-like adaptability and warmth. To fill this gap, we propose the MIND (Multi-agent INner Dialogue), a novel paradigm that provides more immersive psychological healing environments. Considering the strong generative and role-playing ability of LLM agents, we predefine an interactive healing framework and assign LLM agents different roles within the framework to engage in interactive inner dialogues with users, thereby providing an immersive healing experience. We conduct extensive human experiments in various real-world healing dimensions, and find that MIND provides a more user-friendly experience than traditional paradigms. This demonstrates that MIND effectively leverages the significant potential of LLMs in psychological healing.
BASIC: Semi-supervised Multi-organ Segmentation with Balanced Subclass Regularization and Semantic-conflict Penalty
Jan 07, 2025



Abstract:Semi-supervised learning (SSL) has shown notable potential in relieving the heavy demand of dense prediction tasks on large-scale well-annotated datasets, especially for the challenging multi-organ segmentation (MoS). However, the prevailing class-imbalance problem in MoS caused by the substantial variations in organ size exacerbates the learning difficulty of the SSL network. To address this issue, in this paper, we propose an innovative semi-supervised network with BAlanced Subclass regularIzation and semantic-Conflict penalty mechanism (BASIC) to effectively learn the unbiased knowledge for semi-supervised MoS. Concretely, we construct a novel auxiliary subclass segmentation (SCS) task based on priorly generated balanced subclasses, thus deeply excavating the unbiased information for the main MoS task with the fashion of multi-task learning. Additionally, based on a mean teacher framework, we elaborately design a balanced subclass regularization to utilize the teacher predictions of SCS task to supervise the student predictions of MoS task, thus effectively transferring unbiased knowledge to the MoS subnetwork and alleviating the influence of the class-imbalance problem. Considering the similar semantic information inside the subclasses and their corresponding original classes (i.e., parent classes), we devise a semantic-conflict penalty mechanism to give heavier punishments to the conflicting SCS predictions with wrong parent classes and provide a more accurate constraint to the MoS predictions. Extensive experiments conducted on two publicly available datasets, i.e., the WORD dataset and the MICCAI FLARE 2022 dataset, have verified the superior performance of our proposed BASIC compared to other state-of-the-art methods.
BTMuda: A Bi-level Multi-source unsupervised domain adaptation framework for breast cancer diagnosis
Aug 30, 2024Abstract:Deep learning has revolutionized the early detection of breast cancer, resulting in a significant decrease in mortality rates. However, difficulties in obtaining annotations and huge variations in distribution between training sets and real scenes have limited their clinical applications. To address these limitations, unsupervised domain adaptation (UDA) methods have been used to transfer knowledge from one labeled source domain to the unlabeled target domain, yet these approaches suffer from severe domain shift issues and often ignore the potential benefits of leveraging multiple relevant sources in practical applications. To address these limitations, in this work, we construct a Three-Branch Mixed extractor and propose a Bi-level Multi-source unsupervised domain adaptation method called BTMuda for breast cancer diagnosis. Our method addresses the problems of domain shift by dividing domain shift issues into two levels: intra-domain and inter-domain. To reduce the intra-domain shift, we jointly train a CNN and a Transformer as two paths of a domain mixed feature extractor to obtain robust representations rich in both low-level local and high-level global information. As for the inter-domain shift, we redesign the Transformer delicately to a three-branch architecture with cross-attention and distillation, which learns domain-invariant representations from multiple domains. Besides, we introduce two alignment modules - one for feature alignment and one for classifier alignment - to improve the alignment process. Extensive experiments conducted on three public mammographic datasets demonstrate that our BTMuda outperforms state-of-the-art methods.
Alleviating Class Imbalance in Semi-supervised Multi-organ Segmentation via Balanced Subclass Regularization
Aug 26, 2024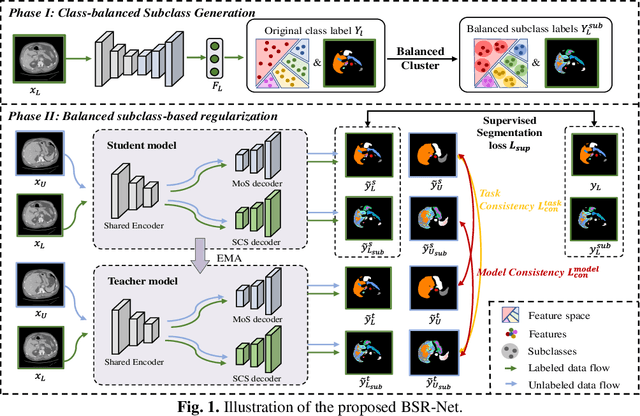
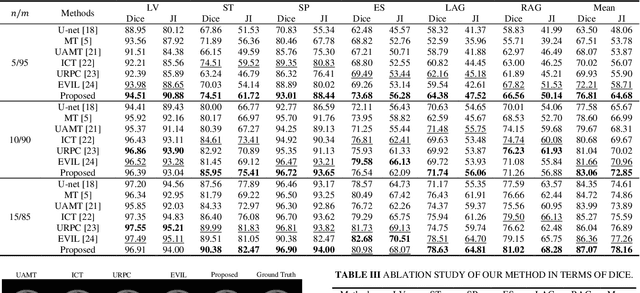
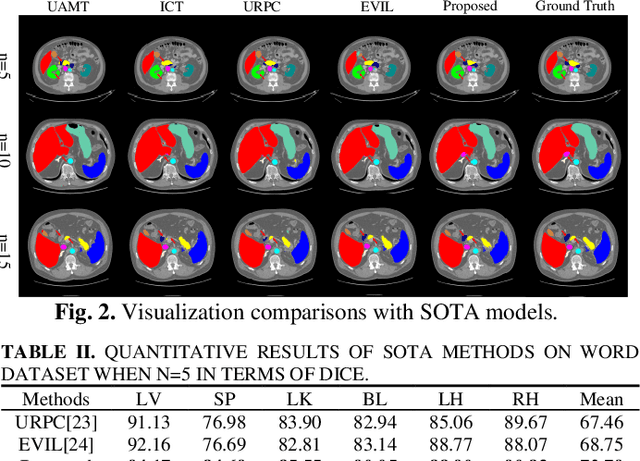
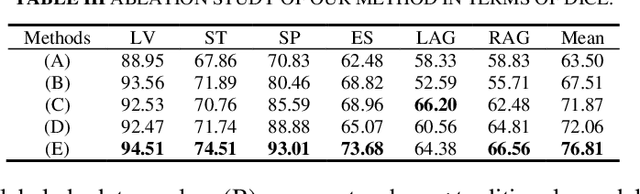
Abstract:Semi-supervised learning (SSL) has shown notable potential in relieving the heavy demand of dense prediction tasks on large-scale well-annotated datasets, especially for the challenging multi-organ segmentation (MoS). However, the prevailing class-imbalance problem in MoS, caused by the substantial variations in organ size, exacerbates the learning difficulty of the SSL network. To alleviate this issue, we present a two-phase semi-supervised network (BSR-Net) with balanced subclass regularization for MoS. Concretely, in Phase I, we introduce a class-balanced subclass generation strategy based on balanced clustering to effectively generate multiple balanced subclasses from original biased ones according to their pixel proportions. Then, in Phase II, we design an auxiliary subclass segmentation (SCS) task within the multi-task framework of the main MoS task. The SCS task contributes a balanced subclass regularization to the main MoS task and transfers unbiased knowledge to the MoS network, thus alleviating the influence of the class-imbalance problem. Extensive experiments conducted on two publicly available datasets, i.e., the MICCAI FLARE 2022 dataset and the WORD dataset, verify the superior performance of our method compared with other methods.
Contrastive Diffusion Model with Auxiliary Guidance for Coarse-to-Fine PET Reconstruction
Aug 20, 2023



Abstract:To obtain high-quality positron emission tomography (PET) scans while reducing radiation exposure to the human body, various approaches have been proposed to reconstruct standard-dose PET (SPET) images from low-dose PET (LPET) images. One widely adopted technique is the generative adversarial networks (GANs), yet recently, diffusion probabilistic models (DPMs) have emerged as a compelling alternative due to their improved sample quality and higher log-likelihood scores compared to GANs. Despite this, DPMs suffer from two major drawbacks in real clinical settings, i.e., the computationally expensive sampling process and the insufficient preservation of correspondence between the conditioning LPET image and the reconstructed PET (RPET) image. To address the above limitations, this paper presents a coarse-to-fine PET reconstruction framework that consists of a coarse prediction module (CPM) and an iterative refinement module (IRM). The CPM generates a coarse PET image via a deterministic process, and the IRM samples the residual iteratively. By delegating most of the computational overhead to the CPM, the overall sampling speed of our method can be significantly improved. Furthermore, two additional strategies, i.e., an auxiliary guidance strategy and a contrastive diffusion strategy, are proposed and integrated into the reconstruction process, which can enhance the correspondence between the LPET image and the RPET image, further improving clinical reliability. Extensive experiments on two human brain PET datasets demonstrate that our method outperforms the state-of-the-art PET reconstruction methods. The source code is available at \url{https://github.com/Show-han/PET-Reconstruction}.
DiffDP: Radiotherapy Dose Prediction via a Diffusion Model
Jul 19, 2023



Abstract:Currently, deep learning (DL) has achieved the automatic prediction of dose distribution in radiotherapy planning, enhancing its efficiency and quality. However, existing methods suffer from the over-smoothing problem for their commonly used L_1 or L_2 loss with posterior average calculations. To alleviate this limitation, we innovatively introduce a diffusion-based dose prediction (DiffDP) model for predicting the radiotherapy dose distribution of cancer patients. Specifically, the DiffDP model contains a forward process and a reverse process. In the forward process, DiffDP gradually transforms dose distribution maps into Gaussian noise by adding small noise and trains a noise predictor to predict the noise added in each timestep. In the reverse process, it removes the noise from the original Gaussian noise in multiple steps with the well-trained noise predictor and finally outputs the predicted dose distribution map. To ensure the accuracy of the prediction, we further design a structure encoder to extract anatomical information from patient anatomy images and enable the noise predictor to be aware of the dose constraints within several essential organs, i.e., the planning target volume and organs at risk. Extensive experiments on an in-house dataset with 130 rectum cancer patients demonstrate the s
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge