Pinxian Zeng
BTMuda: A Bi-level Multi-source unsupervised domain adaptation framework for breast cancer diagnosis
Aug 30, 2024Abstract:Deep learning has revolutionized the early detection of breast cancer, resulting in a significant decrease in mortality rates. However, difficulties in obtaining annotations and huge variations in distribution between training sets and real scenes have limited their clinical applications. To address these limitations, unsupervised domain adaptation (UDA) methods have been used to transfer knowledge from one labeled source domain to the unlabeled target domain, yet these approaches suffer from severe domain shift issues and often ignore the potential benefits of leveraging multiple relevant sources in practical applications. To address these limitations, in this work, we construct a Three-Branch Mixed extractor and propose a Bi-level Multi-source unsupervised domain adaptation method called BTMuda for breast cancer diagnosis. Our method addresses the problems of domain shift by dividing domain shift issues into two levels: intra-domain and inter-domain. To reduce the intra-domain shift, we jointly train a CNN and a Transformer as two paths of a domain mixed feature extractor to obtain robust representations rich in both low-level local and high-level global information. As for the inter-domain shift, we redesign the Transformer delicately to a three-branch architecture with cross-attention and distillation, which learns domain-invariant representations from multiple domains. Besides, we introduce two alignment modules - one for feature alignment and one for classifier alignment - to improve the alignment process. Extensive experiments conducted on three public mammographic datasets demonstrate that our BTMuda outperforms state-of-the-art methods.
S3PET: Semi-supervised Standard-dose PET Image Reconstruction via Dose-aware Token Swap
Jul 30, 2024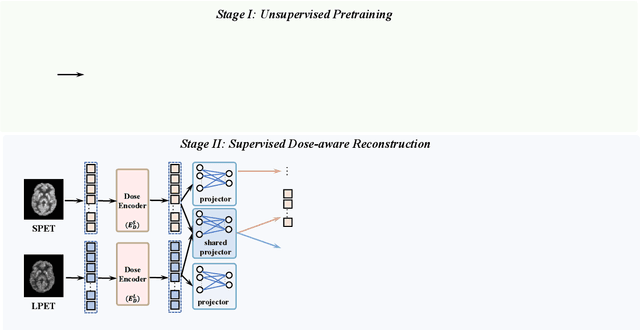
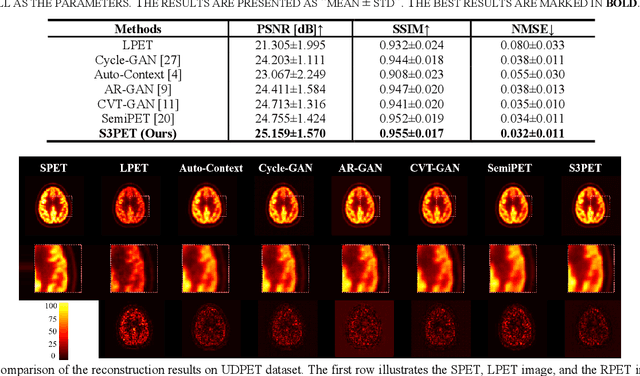
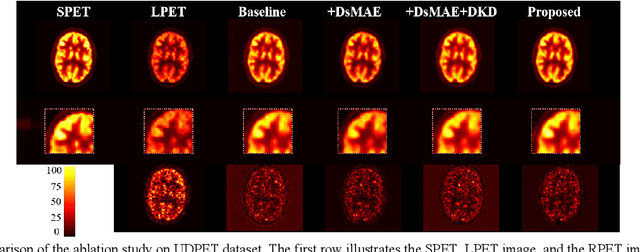
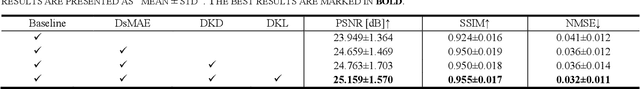
Abstract:To acquire high-quality positron emission tomography (PET) images while reducing the radiation tracer dose, numerous efforts have been devoted to reconstructing standard-dose PET (SPET) images from low-dose PET (LPET). However, the success of current fully-supervised approaches relies on abundant paired LPET and SPET images, which are often unavailable in clinic. Moreover, these methods often mix the dose-invariant content with dose level-related dose-specific details during reconstruction, resulting in distorted images. To alleviate these problems, in this paper, we propose a two-stage Semi-Supervised SPET reconstruction framework, namely S3PET, to accommodate the training of abundant unpaired and limited paired SPET and LPET images. Our S3PET involves an un-supervised pre-training stage (Stage I) to extract representations from unpaired images, and a supervised dose-aware reconstruction stage (Stage II) to achieve LPET-to-SPET reconstruction by transferring the dose-specific knowledge between paired images. Specifically, in stage I, two independent dose-specific masked autoencoders (DsMAEs) are adopted to comprehensively understand the unpaired SPET and LPET images. Then, in Stage II, the pre-trained DsMAEs are further finetuned using paired images. To prevent distortions in both content and details, we introduce two elaborate modules, i.e., a dose knowledge decouple module to disentangle the respective dose-specific and dose-invariant knowledge of LPET and SPET, and a dose-specific knowledge learning module to transfer the dose-specific information from SPET to LPET, thereby achieving high-quality SPET reconstruction from LPET images. Experiments on two datasets demonstrate that our S3PET achieves state-of-the-art performance quantitatively and qualitatively.
MCAD: Multi-modal Conditioned Adversarial Diffusion Model for High-Quality PET Image Reconstruction
Jun 19, 2024Abstract:Radiation hazards associated with standard-dose positron emission tomography (SPET) images remain a concern, whereas the quality of low-dose PET (LPET) images fails to meet clinical requirements. Therefore, there is great interest in reconstructing SPET images from LPET images. However, prior studies focus solely on image data, neglecting vital complementary information from other modalities, e.g., patients' clinical tabular, resulting in compromised reconstruction with limited diagnostic utility. Moreover, they often overlook the semantic consistency between real SPET and reconstructed images, leading to distorted semantic contexts. To tackle these problems, we propose a novel Multi-modal Conditioned Adversarial Diffusion model (MCAD) to reconstruct SPET images from multi-modal inputs, including LPET images and clinical tabular. Specifically, our MCAD incorporates a Multi-modal conditional Encoder (Mc-Encoder) to extract multi-modal features, followed by a conditional diffusion process to blend noise with multi-modal features and gradually map blended features to the target SPET images. To balance multi-modal inputs, the Mc-Encoder embeds Optimal Multi-modal Transport co-Attention (OMTA) to narrow the heterogeneity gap between image and tabular while capturing their interactions, providing sufficient guidance for reconstruction. In addition, to mitigate semantic distortions, we introduce the Multi-Modal Masked Text Reconstruction (M3TRec), which leverages semantic knowledge extracted from denoised PET images to restore the masked clinical tabular, thereby compelling the network to maintain accurate semantics during reconstruction. To expedite the diffusion process, we further introduce an adversarial diffusive network with a reduced number of diffusion steps. Experiments show that our method achieves the state-of-the-art performance both qualitatively and quantitatively.
Image2Points:A 3D Point-based Context Clusters GAN for High-Quality PET Image Reconstruction
Feb 01, 2024Abstract:To obtain high-quality Positron emission tomography (PET) images while minimizing radiation exposure, numerous methods have been proposed to reconstruct standard-dose PET (SPET) images from the corresponding low-dose PET (LPET) images. However, these methods heavily rely on voxel-based representations, which fall short of adequately accounting for the precise structure and fine-grained context, leading to compromised reconstruction. In this paper, we propose a 3D point-based context clusters GAN, namely PCC-GAN, to reconstruct high-quality SPET images from LPET. Specifically, inspired by the geometric representation power of points, we resort to a point-based representation to enhance the explicit expression of the image structure, thus facilitating the reconstruction with finer details. Moreover, a context clustering strategy is applied to explore the contextual relationships among points, which mitigates the ambiguities of small structures in the reconstructed images. Experiments on both clinical and phantom datasets demonstrate that our PCC-GAN outperforms the state-of-the-art reconstruction methods qualitatively and quantitatively. Code is available at https://github.com/gluucose/PCCGAN.
TriDo-Former: A Triple-Domain Transformer for Direct PET Reconstruction from Low-Dose Sinograms
Aug 10, 2023Abstract:To obtain high-quality positron emission tomography (PET) images while minimizing radiation exposure, various methods have been proposed for reconstructing standard-dose PET (SPET) images from low-dose PET (LPET) sinograms directly. However, current methods often neglect boundaries during sinogram-to-image reconstruction, resulting in high-frequency distortion in the frequency domain and diminished or fuzzy edges in the reconstructed images. Furthermore, the convolutional architectures, which are commonly used, lack the ability to model long-range non-local interactions, potentially leading to inaccurate representations of global structures. To alleviate these problems, we propose a transformer-based model that unites triple domains of sinogram, image, and frequency for direct PET reconstruction, namely TriDo-Former. Specifically, the TriDo-Former consists of two cascaded networks, i.e., a sinogram enhancement transformer (SE-Former) for denoising the input LPET sinograms and a spatial-spectral reconstruction transformer (SSR-Former) for reconstructing SPET images from the denoised sinograms. Different from the vanilla transformer that splits an image into 2D patches, based specifically on the PET imaging mechanism, our SE-Former divides the sinogram into 1D projection view angles to maintain its inner-structure while denoising, preventing the noise in the sinogram from prorogating into the image domain. Moreover, to mitigate high-frequency distortion and improve reconstruction details, we integrate global frequency parsers (GFPs) into SSR-Former. The GFP serves as a learnable frequency filter that globally adjusts the frequency components in the frequency domain, enforcing the network to restore high-frequency details resembling real SPET images. Validations on a clinical dataset demonstrate that our TriDo-Former outperforms the state-of-the-art methods qualitatively and quantitatively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge