Jianghong Xiao
FDDM: Frequency-Decomposed Diffusion Model for Rectum Cancer Dose Prediction in Radiotherapy
Oct 10, 2024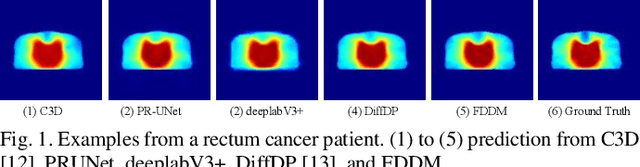
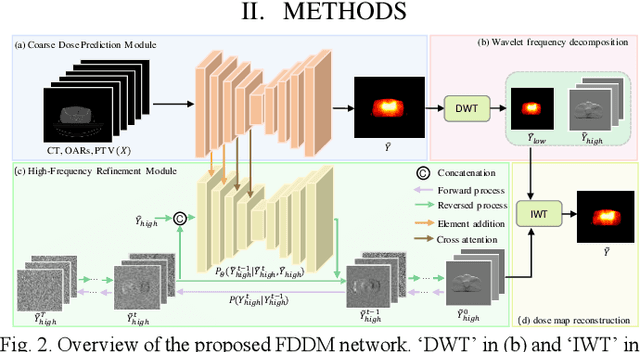
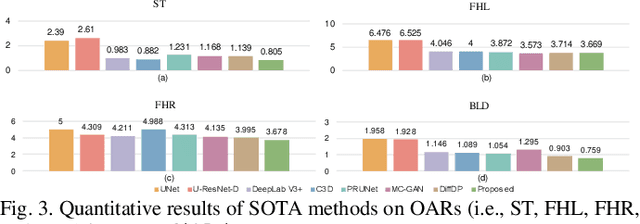
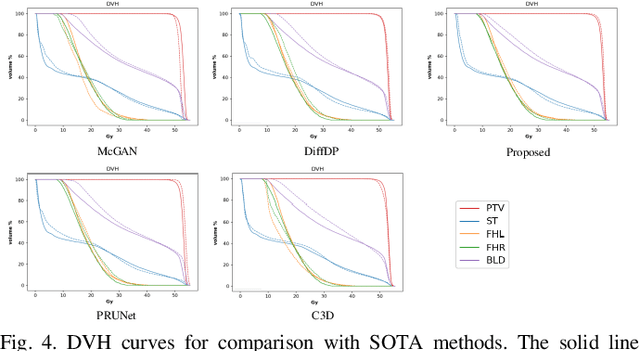
Abstract:Accurate dose distribution prediction is crucial in the radiotherapy planning. Although previous methods based on convolutional neural network have shown promising performance, they have the problem of over-smoothing, leading to prediction without important high-frequency details. Recently, diffusion model has achieved great success in computer vision, which excels in generating images with more high-frequency details, yet suffers from time-consuming and extensive computational resource consumption. To alleviate these problems, we propose Frequency-Decomposed Diffusion Model (FDDM) that refines the high-frequency subbands of the dose map. To be specific, we design a Coarse Dose Prediction Module (CDPM) to first predict a coarse dose map and then utilize discrete wavelet transform to decompose the coarse dose map into a low-frequency subband and three high?frequency subbands. There is a notable difference between the coarse predicted results and ground truth in high?frequency subbands. Therefore, we design a diffusion-based module called High-Frequency Refinement Module (HFRM) that performs diffusion operation in the high?frequency components of the dose map instead of the original dose map. Extensive experiments on an in-house dataset verify the effectiveness of our approach.
Dose Prediction Driven Radiotherapy Paramters Regression via Intra- and Inter-Relation Modeling
Feb 29, 2024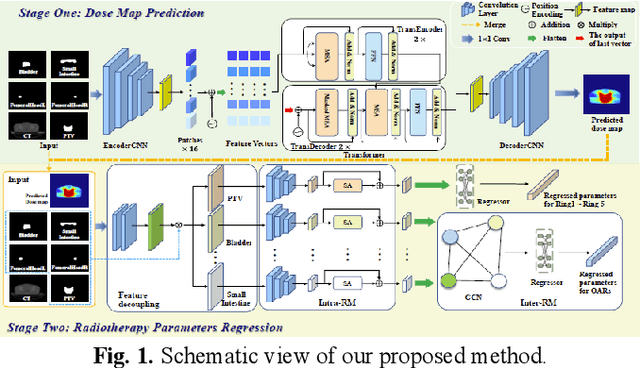
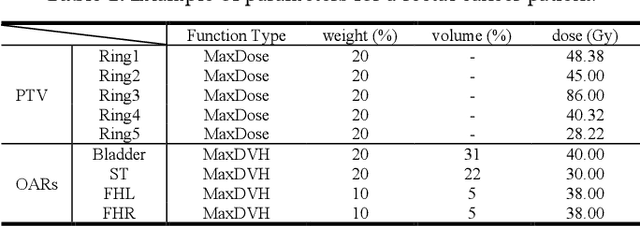
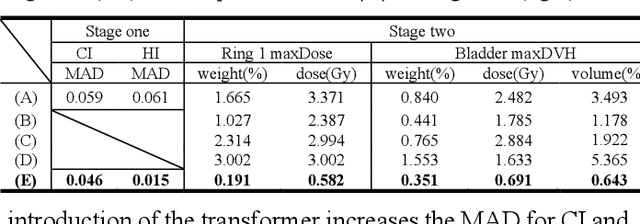

Abstract:Deep learning has facilitated the automation of radiotherapy by predicting accurate dose distribution maps. However, existing methods fail to derive the desirable radiotherapy parameters that can be directly input into the treatment planning system (TPS), impeding the full automation of radiotherapy. To enable more thorough automatic radiotherapy, in this paper, we propose a novel two-stage framework to directly regress the radiotherapy parameters, including a dose map prediction stage and a radiotherapy parameters regression stage. In stage one, we combine transformer and convolutional neural network (CNN) to predict realistic dose maps with rich global and local information, providing accurate dosimetric knowledge for the subsequent parameters regression. In stage two, two elaborate modules, i.e., an intra-relation modeling (Intra-RM) module and an inter-relation modeling (Inter-RM) module, are designed to exploit the organ-specific and organ-shared features for precise parameters regression. Experimental results on a rectal cancer dataset demonstrate the effectiveness of our method.
Diffusion-based Radiotherapy Dose Prediction Guided by Inter-slice Aware Structure Encoding
Nov 06, 2023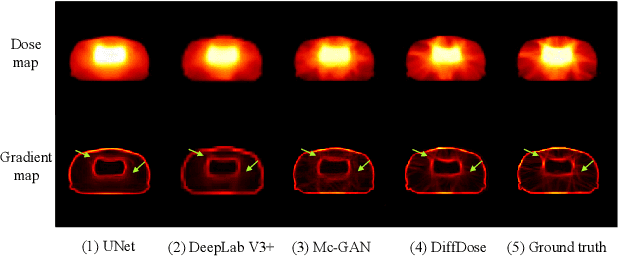
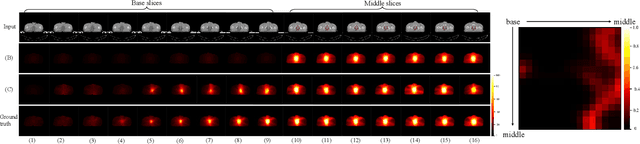
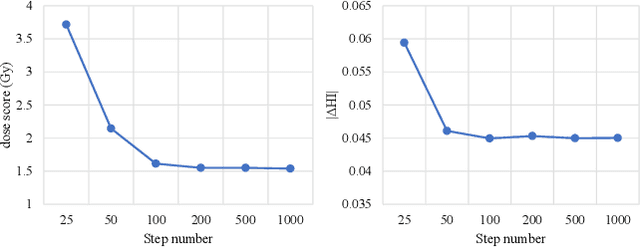

Abstract:Deep learning (DL) has successfully automated dose distribution prediction in radiotherapy planning, enhancing both efficiency and quality. However, existing methods suffer from the over-smoothing problem for their commonly used L1 or L2 loss with posterior average calculations. To alleviate this limitation, we propose a diffusion model-based method (DiffDose) for predicting the radiotherapy dose distribution of cancer patients. Specifically, the DiffDose model contains a forward process and a reverse process. In the forward process, DiffDose transforms dose distribution maps into pure Gaussian noise by gradually adding small noise and a noise predictor is simultaneously trained to estimate the noise added at each timestep. In the reverse process, it removes the noise from the pure Gaussian noise in multiple steps with the well-trained noise predictor and finally outputs the predicted dose distribution maps...
Polymerized Feature-based Domain Adaptation for Cervical Cancer Dose Map Prediction
Aug 20, 2023



Abstract:Recently, deep learning (DL) has automated and accelerated the clinical radiation therapy (RT) planning significantly by predicting accurate dose maps. However, most DL-based dose map prediction methods are data-driven and not applicable for cervical cancer where only a small amount of data is available. To address this problem, this paper proposes to transfer the rich knowledge learned from another cancer, i.e., rectum cancer, which has the same scanning area and more clinically available data, to improve the dose map prediction performance for cervical cancer through domain adaptation. In order to close the congenital domain gap between the source (i.e., rectum cancer) and the target (i.e., cervical cancer) domains, we develop an effective Transformer-based polymerized feature module (PFM), which can generate an optimal polymerized feature distribution to smoothly align the two input distributions. Experimental results on two in-house clinical datasets demonstrate the superiority of the proposed method compared with state-of-the-art methods.
WORD: Revisiting Organs Segmentation in the Whole Abdominal Region
Nov 17, 2021



Abstract:Whole abdominal organs segmentation plays an important role in abdomen lesion diagnosis, radiotherapy planning, and follow-up. However, delineating all abdominal organs by oncologists manually is time-consuming and very expensive. Recently, deep learning-based medical image segmentation has shown the potential to reduce manual delineation efforts, but it still requires a large-scale fine annotated dataset for training. Although many efforts in this task, there are still few large image datasets covering the whole abdomen region with accurate and detailed annotations for the whole abdominal organ segmentation. In this work, we establish a large-scale \textit{W}hole abdominal \textit{OR}gans \textit{D}ataset (\textit{WORD}) for algorithms research and clinical applications development. This dataset contains 150 abdominal CT volumes (30495 slices) and each volume has 16 organs with fine pixel-level annotations and scribble-based sparse annotation, which may be the largest dataset with whole abdominal organs annotation. Several state-of-the-art segmentation methods are evaluated on this dataset. And, we also invited clinical oncologists to revise the model predictions to measure the gap between the deep learning method and real oncologists. We further introduce and evaluate a new scribble-based weakly supervised segmentation on this dataset. The work provided a new benchmark for the abdominal multi-organ segmentation task and these experiments can serve as the baseline for future research and clinical application development. The codebase and dataset will be released at: https://github.com/HiLab-git/WORD
DSU-net: Dense SegU-net for automatic head-and-neck tumor segmentation in MR images
Jun 12, 2020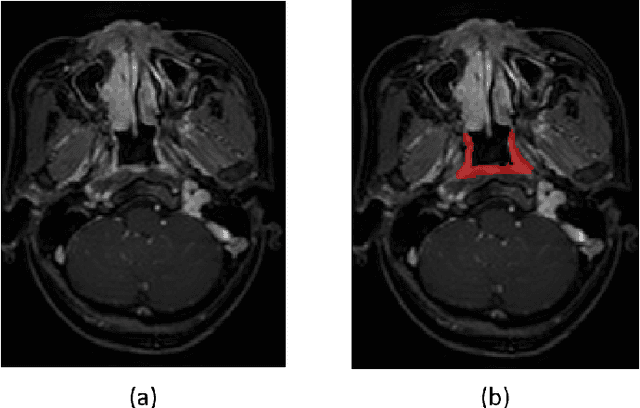
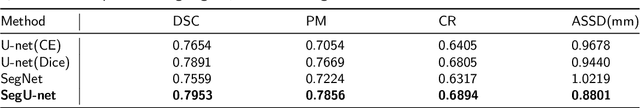
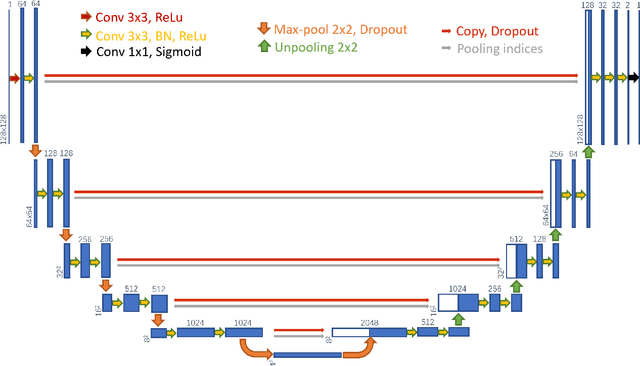
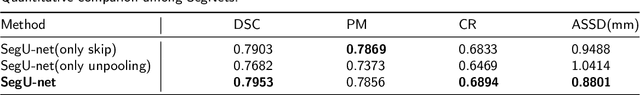
Abstract:Precise and accurate segmentation of the most common head-and-neck tumor, nasopharyngeal carcinoma (NPC), in MRI sheds light on treatment and regulatory decisions making. However, the large variations in the lesion size and shape of NPC, boundary ambiguity, as well as the limited available annotated samples conspire NPC segmentation in MRI towards a challenging task. In this paper, we propose a Dense SegU-net (DSU-net) framework for automatic NPC segmentation in MRI. Our contribution is threefold. First, different from the traditional decoder in U-net using upconvolution for upsamling, we argue that the restoration from low resolution features to high resolution output should be capable of preserving information significant for precise boundary localization. Hence, we use unpooling to unsample and propose SegU-net. Second, to combat the potential vanishing-gradient problem, we introduce dense blocks which can facilitate feature propagation and reuse. Third, using only cross entropy (CE) as loss function may bring about troubles such as miss-prediction, therefore we propose to use a loss function comprised of both CE loss and Dice loss to train the network. Quantitative and qualitative comparisons are carried out extensively on in-house datasets, the experimental results show that our proposed architecture outperforms the existing state-of-the-art segmentation networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge