Mei Hong
Alleviating neighbor bias: augmenting graph self-supervise learning with structural equivalent positive samples
Dec 08, 2022Abstract:In recent years, using a self-supervised learning framework to learn the general characteristics of graphs has been considered a promising paradigm for graph representation learning. The core of self-supervised learning strategies for graph neural networks lies in constructing suitable positive sample selection strategies. However, existing GNNs typically aggregate information from neighboring nodes to update node representations, leading to an over-reliance on neighboring positive samples, i.e., homophilous samples; while ignoring long-range positive samples, i.e., positive samples that are far apart on the graph but structurally equivalent samples, a problem we call "neighbor bias." This neighbor bias can reduce the generalization performance of GNNs. In this paper, we argue that the generalization properties of GNNs should be determined by combining homogeneous samples and structurally equivalent samples, which we call the "GC combination hypothesis." Therefore, we propose a topological signal-driven self-supervised method. It uses a topological information-guided structural equivalence sampling strategy. First, we extract multiscale topological features using persistent homology. Then we compute the structural equivalence of node pairs based on their topological features. In particular, we design a topological loss function to pull in non-neighboring node pairs with high structural equivalence in the representation space to alleviate neighbor bias. Finally, we use the joint training mechanism to adjust the effect of structural equivalence on the model to fit datasets with different characteristics. We conducted experiments on the node classification task across seven graph datasets. The results show that the model performance can be effectively improved using a strategy of topological signal enhancement.
Deep Learning based Multi-modal Computing with Feature Disentanglement for MRI Image Synthesis
May 06, 2021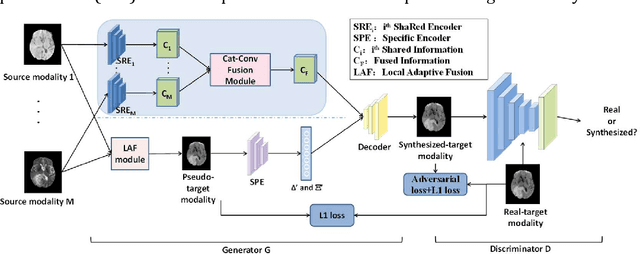
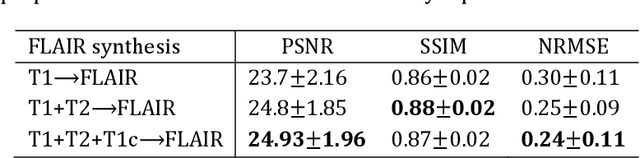
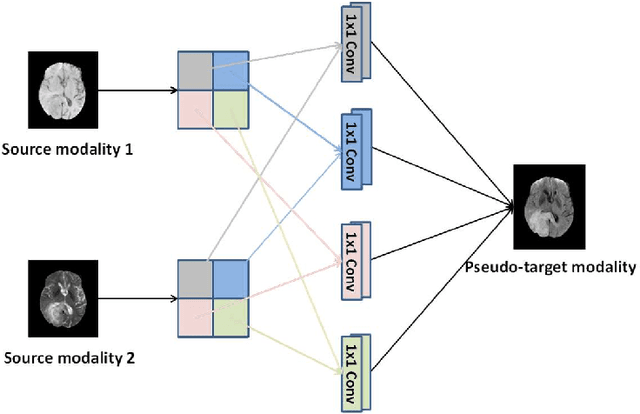
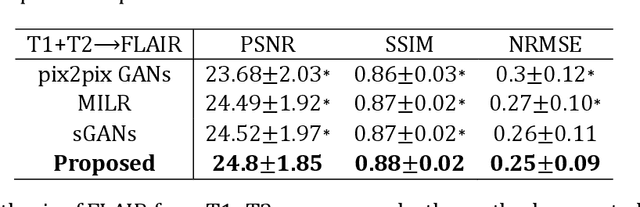
Abstract:Purpose: Different Magnetic resonance imaging (MRI) modalities of the same anatomical structure are required to present different pathological information from the physical level for diagnostic needs. However, it is often difficult to obtain full-sequence MRI images of patients owing to limitations such as time consumption and high cost. The purpose of this work is to develop an algorithm for target MRI sequences prediction with high accuracy, and provide more information for clinical diagnosis. Methods: We propose a deep learning based multi-modal computing model for MRI synthesis with feature disentanglement strategy. To take full advantage of the complementary information provided by different modalities, multi-modal MRI sequences are utilized as input. Notably, the proposed approach decomposes each input modality into modality-invariant space with shared information and modality-specific space with specific information, so that features are extracted separately to effectively process the input data. Subsequently, both of them are fused through the adaptive instance normalization (AdaIN) layer in the decoder. In addition, to address the lack of specific information of the target modality in the test phase, a local adaptive fusion (LAF) module is adopted to generate a modality-like pseudo-target with specific information similar to the ground truth. Results: To evaluate the synthesis performance, we verify our method on the BRATS2015 dataset of 164 subjects. The experimental results demonstrate our approach significantly outperforms the benchmark method and other state-of-the-art medical image synthesis methods in both quantitative and qualitative measures. Compared with the pix2pixGANs method, the PSNR improves from 23.68 to 24.8. Conclusion: The proposed method could be effective in prediction of target MRI sequences, and useful for clinical diagnosis and treatment.
ASMFS: Adaptive-Similarity-based Multi-modality Feature Selection for Classification of Alzheimer's Disease
Oct 16, 2020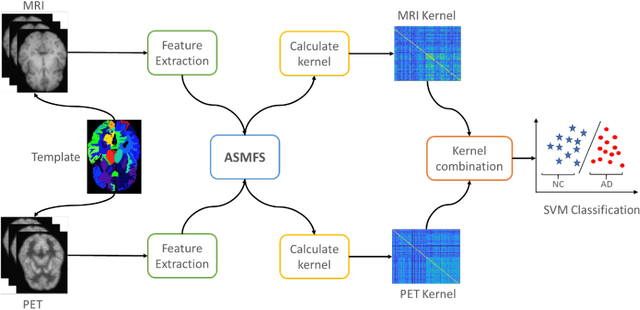
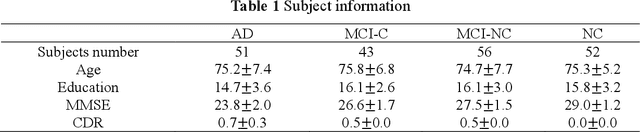
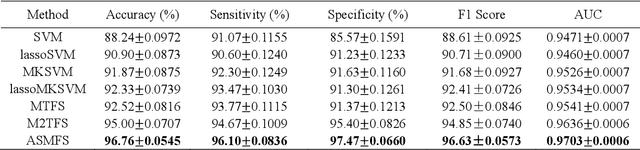
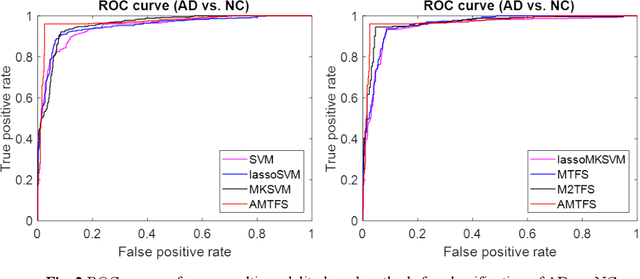
Abstract:With the increasing amounts of high-dimensional heterogeneous data to be processed, multi-modality feature selection has become an important research direction in medical image analysis. Traditional methods usually depict the data structure using fixed and predefined similarity matrix for each modality separately, without considering the potential relationship structure across different modalities. In this paper, we propose a novel multi-modality feature selection method, which performs feature selection and local similarity learning simultaniously. Specially, a similarity matrix is learned by jointly considering different imaging modalities. And at the same time, feature selection is conducted by imposing sparse l_{2, 1} norm constraint. The effectiveness of our proposed joint learning method can be well demonstrated by the experimental results on Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset, which outperforms existing the state-of-the-art multi-modality approaches.
DSU-net: Dense SegU-net for automatic head-and-neck tumor segmentation in MR images
Jun 12, 2020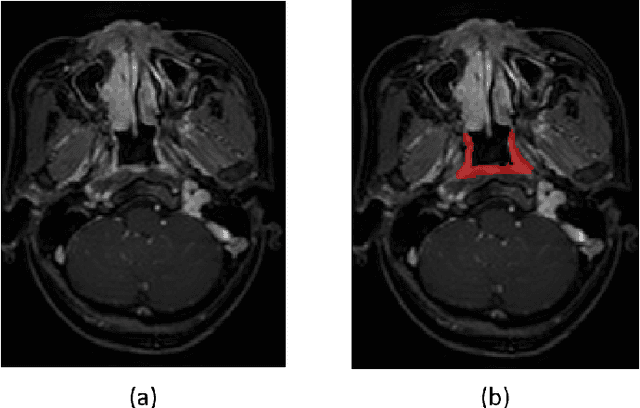
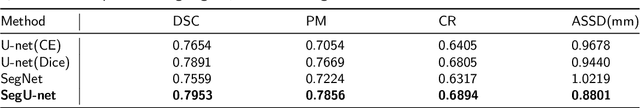
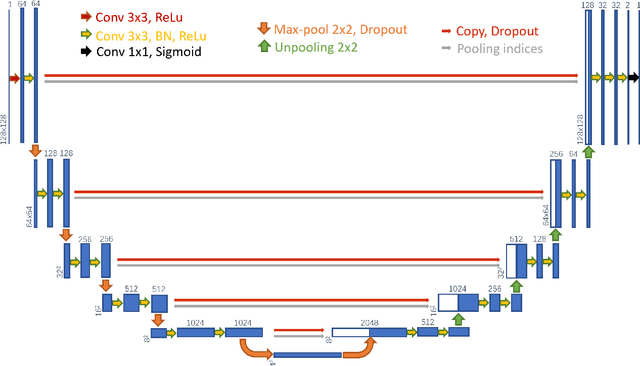
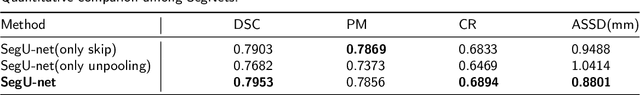
Abstract:Precise and accurate segmentation of the most common head-and-neck tumor, nasopharyngeal carcinoma (NPC), in MRI sheds light on treatment and regulatory decisions making. However, the large variations in the lesion size and shape of NPC, boundary ambiguity, as well as the limited available annotated samples conspire NPC segmentation in MRI towards a challenging task. In this paper, we propose a Dense SegU-net (DSU-net) framework for automatic NPC segmentation in MRI. Our contribution is threefold. First, different from the traditional decoder in U-net using upconvolution for upsamling, we argue that the restoration from low resolution features to high resolution output should be capable of preserving information significant for precise boundary localization. Hence, we use unpooling to unsample and propose SegU-net. Second, to combat the potential vanishing-gradient problem, we introduce dense blocks which can facilitate feature propagation and reuse. Third, using only cross entropy (CE) as loss function may bring about troubles such as miss-prediction, therefore we propose to use a loss function comprised of both CE loss and Dice loss to train the network. Quantitative and qualitative comparisons are carried out extensively on in-house datasets, the experimental results show that our proposed architecture outperforms the existing state-of-the-art segmentation networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge