Pietro Lio
Molecular Representations in Implicit Functional Space via Hyper-Networks
Jan 29, 2026Abstract:Molecular representations fundamentally shape how machine learning systems reason about molecular structure and physical properties. Most existing approaches adopt a discrete pipeline: molecules are encoded as sequences, graphs, or point clouds, mapped to fixed-dimensional embeddings, and then used for task-specific prediction. This paradigm treats molecules as discrete objects, despite their intrinsically continuous and field-like physical nature. We argue that molecular learning can instead be formulated as learning in function space. Specifically, we model each molecule as a continuous function over three-dimensional (3D) space and treat this molecular field as the primary object of representation. From this perspective, conventional molecular representations arise as particular sampling schemes of an underlying continuous object. We instantiate this formulation with MolField, a hyper-network-based framework that learns distributions over molecular fields. To ensure physical consistency, these functions are defined over canonicalized coordinates, yielding invariance to global SE(3) transformations. To enable learning directly over functions, we introduce a structured weight tokenization and train a sequence-based hyper-network to model a shared prior over molecular fields. We evaluate MolField on molecular dynamics and property prediction. Our results show that treating molecules as continuous functions fundamentally changes how molecular representations generalize across tasks and yields downstream behavior that is stable to how molecules are discretized or queried.
A Monosemantic Attribution Framework for Stable Interpretability in Clinical Neuroscience Large Language Models
Jan 25, 2026Abstract:Interpretability remains a key challenge for deploying large language models (LLMs) in clinical settings such as Alzheimer's disease progression diagnosis, where early and trustworthy predictions are essential. Existing attribution methods exhibit high inter-method variability and unstable explanations due to the polysemantic nature of LLM representations, while mechanistic interpretability approaches lack direct alignment with model inputs and outputs and do not provide explicit importance scores. We introduce a unified interpretability framework that integrates attributional and mechanistic perspectives through monosemantic feature extraction. By constructing a monosemantic embedding space at the level of an LLM layer and optimizing the framework to explicitly reduce inter-method variability, our approach produces stable input-level importance scores and highlights salient features via a decompressed representation of the layer of interest, advancing the safe and trustworthy application of LLMs in cognitive health and neurodegenerative disease.
MODE: Efficient Time Series Prediction with Mamba Enhanced by Low-Rank Neural ODEs
Jan 01, 2026Abstract:Time series prediction plays a pivotal role across diverse domains such as finance, healthcare, energy systems, and environmental modeling. However, existing approaches often struggle to balance efficiency, scalability, and accuracy, particularly when handling long-range dependencies and irregularly sampled data. To address these challenges, we propose MODE, a unified framework that integrates Low-Rank Neural Ordinary Differential Equations (Neural ODEs) with an Enhanced Mamba architecture. As illustrated in our framework, the input sequence is first transformed by a Linear Tokenization Layer and then processed through multiple Mamba Encoder blocks, each equipped with an Enhanced Mamba Layer that employs Causal Convolution, SiLU activation, and a Low-Rank Neural ODE enhancement to efficiently capture temporal dynamics. This low-rank formulation reduces computational overhead while maintaining expressive power. Furthermore, a segmented selective scanning mechanism, inspired by pseudo-ODE dynamics, adaptively focuses on salient subsequences to improve scalability and long-range sequence modeling. Extensive experiments on benchmark datasets demonstrate that MODE surpasses existing baselines in both predictive accuracy and computational efficiency. Overall, our contributions include: (1) a unified and efficient architecture for long-term time series modeling, (2) integration of Mamba's selective scanning with low-rank Neural ODEs for enhanced temporal representation, and (3) substantial improvements in efficiency and scalability enabled by low-rank approximation and dynamic selective scanning.
Deep Spectral Prior
May 26, 2025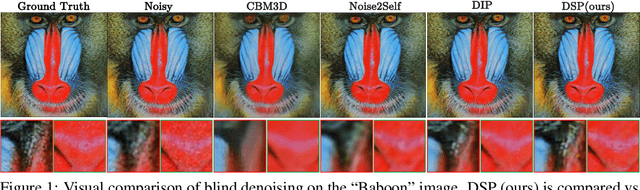
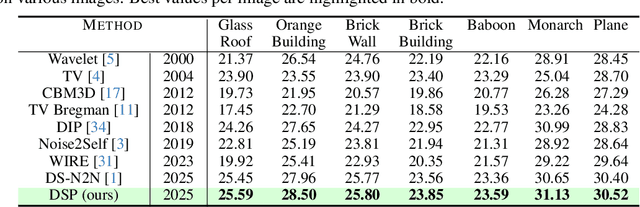


Abstract:We introduce Deep Spectral Prior (DSP), a new formulation of Deep Image Prior (DIP) that redefines image reconstruction as a frequency-domain alignment problem. Unlike traditional DIP, which relies on pixel-wise loss and early stopping to mitigate overfitting, DSP directly matches Fourier coefficients between the network output and observed measurements. This shift introduces an explicit inductive bias towards spectral coherence, aligning with the known frequency structure of images and the spectral bias of convolutional neural networks. We provide a rigorous theoretical framework demonstrating that DSP acts as an implicit spectral regulariser, suppressing high-frequency noise by design and eliminating the need for early stopping. Our analysis spans four core dimensions establishing smooth convergence dynamics, local stability, and favourable bias-variance tradeoffs. We further show that DSP naturally projects reconstructions onto a frequency-consistent manifold, enhancing interpretability and robustness. These theoretical guarantees are supported by empirical results across denoising, inpainting, and super-resolution tasks, where DSP consistently outperforms classical DIP and other unsupervised baselines.
How Particle System Theory Enhances Hypergraph Message Passing
May 24, 2025Abstract:Hypergraphs effectively model higher-order relationships in natural phenomena, capturing complex interactions beyond pairwise connections. We introduce a novel hypergraph message passing framework inspired by interacting particle systems, where hyperedges act as fields inducing shared node dynamics. By incorporating attraction, repulsion, and Allen-Cahn forcing terms, particles of varying classes and features achieve class-dependent equilibrium, enabling separability through the particle-driven message passing. We investigate both first-order and second-order particle system equations for modeling these dynamics, which mitigate over-smoothing and heterophily thus can capture complete interactions. The more stable second-order system permits deeper message passing. Furthermore, we enhance deterministic message passing with stochastic element to account for interaction uncertainties. We prove theoretically that our approach mitigates over-smoothing by maintaining a positive lower bound on the hypergraph Dirichlet energy during propagation and thus to enable hypergraph message passing to go deep. Empirically, our models demonstrate competitive performance on diverse real-world hypergraph node classification tasks, excelling on both homophilic and heterophilic datasets.
Graph Foundation Models: A Comprehensive Survey
May 21, 2025Abstract:Graph-structured data pervades domains such as social networks, biological systems, knowledge graphs, and recommender systems. While foundation models have transformed natural language processing, vision, and multimodal learning through large-scale pretraining and generalization, extending these capabilities to graphs -- characterized by non-Euclidean structures and complex relational semantics -- poses unique challenges and opens new opportunities. To this end, Graph Foundation Models (GFMs) aim to bring scalable, general-purpose intelligence to structured data, enabling broad transfer across graph-centric tasks and domains. This survey provides a comprehensive overview of GFMs, unifying diverse efforts under a modular framework comprising three key components: backbone architectures, pretraining strategies, and adaptation mechanisms. We categorize GFMs by their generalization scope -- universal, task-specific, and domain-specific -- and review representative methods, key innovations, and theoretical insights within each category. Beyond methodology, we examine theoretical foundations including transferability and emergent capabilities, and highlight key challenges such as structural alignment, heterogeneity, scalability, and evaluation. Positioned at the intersection of graph learning and general-purpose AI, GFMs are poised to become foundational infrastructure for open-ended reasoning over structured data. This survey consolidates current progress and outlines future directions to guide research in this rapidly evolving field. Resources are available at https://github.com/Zehong-Wang/Awesome-Foundation-Models-on-Graphs.
Multi-Level Monte Carlo Training of Neural Operators
May 19, 2025Abstract:Operator learning is a rapidly growing field that aims to approximate nonlinear operators related to partial differential equations (PDEs) using neural operators. These rely on discretization of input and output functions and are, usually, expensive to train for large-scale problems at high-resolution. Motivated by this, we present a Multi-Level Monte Carlo (MLMC) approach to train neural operators by leveraging a hierarchy of resolutions of function dicretization. Our framework relies on using gradient corrections from fewer samples of fine-resolution data to decrease the computational cost of training while maintaining a high level accuracy. The proposed MLMC training procedure can be applied to any architecture accepting multi-resolution data. Our numerical experiments on a range of state-of-the-art models and test-cases demonstrate improved computational efficiency compared to traditional single-resolution training approaches, and highlight the existence of a Pareto curve between accuracy and computational time, related to the number of samples per resolution.
Hierarchical Planning for Complex Tasks with Knowledge Graph-RAG and Symbolic Verification
Apr 06, 2025Abstract:Large Language Models (LLMs) have shown promise as robotic planners but often struggle with long-horizon and complex tasks, especially in specialized environments requiring external knowledge. While hierarchical planning and Retrieval-Augmented Generation (RAG) address some of these challenges, they remain insufficient on their own and a deeper integration is required for achieving more reliable systems. To this end, we propose a neuro-symbolic approach that enhances LLMs-based planners with Knowledge Graph-based RAG for hierarchical plan generation. This method decomposes complex tasks into manageable subtasks, further expanded into executable atomic action sequences. To ensure formal correctness and proper decomposition, we integrate a Symbolic Validator, which also functions as a failure detector by aligning expected and observed world states. Our evaluation against baseline methods demonstrates the consistent significant advantages of integrating hierarchical planning, symbolic verification, and RAG across tasks of varying complexity and different LLMs. Additionally, our experimental setup and novel metrics not only validate our approach for complex planning but also serve as a tool for assessing LLMs' reasoning and compositional capabilities.
Debiasing Guidance for Discrete Diffusion with Sequential Monte Carlo
Feb 10, 2025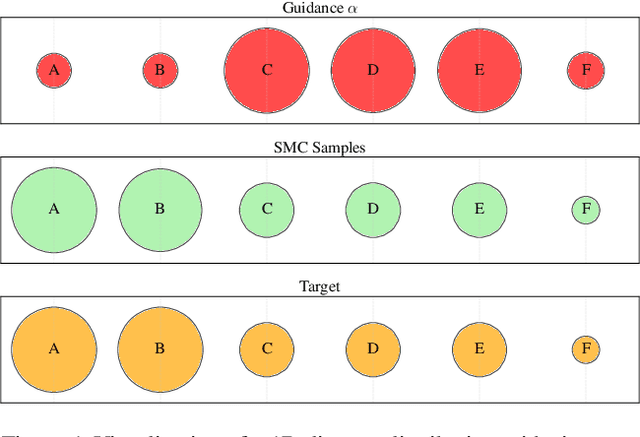
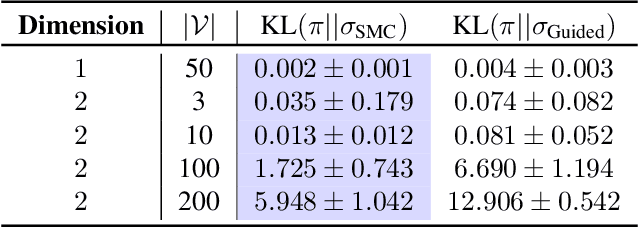
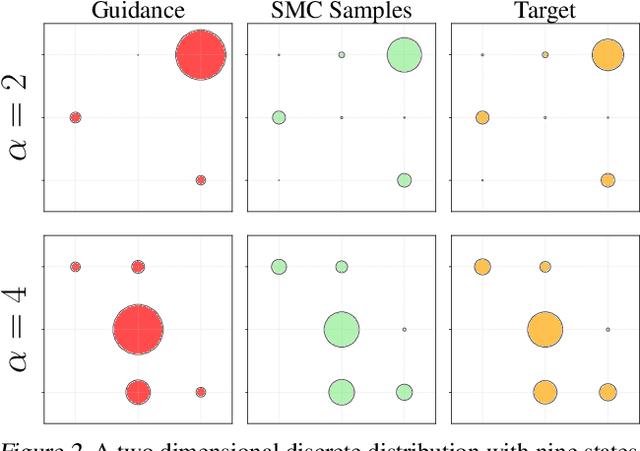

Abstract:Discrete diffusion models are a class of generative models that produce samples from an approximated data distribution within a discrete state space. Often, there is a need to target specific regions of the data distribution. Current guidance methods aim to sample from a distribution with mass proportional to $p_0(x_0) p(\zeta|x_0)^\alpha$ but fail to achieve this in practice. We introduce a Sequential Monte Carlo algorithm that generates unbiasedly from this target distribution, utilising the learnt unconditional and guided process. We validate our approach on low-dimensional distributions, controlled images and text generations. For text generation, our method provides strong control while maintaining low perplexity compared to guidance-based approaches.
KG4Diagnosis: A Hierarchical Multi-Agent LLM Framework with Knowledge Graph Enhancement for Medical Diagnosis
Dec 22, 2024Abstract:Integrating Large Language Models (LLMs) in healthcare diagnosis demands systematic frameworks that can handle complex medical scenarios while maintaining specialized expertise. We present KG4Diagnosis, a novel hierarchical multi-agent framework that combines LLMs with automated knowledge graph construction, encompassing 362 common diseases across medical specialties. Our framework mirrors real-world medical systems through a two-tier architecture: a general practitioner (GP) agent for initial assessment and triage, coordinating with specialized agents for in-depth diagnosis in specific domains. The core innovation lies in our end-to-end knowledge graph generation methodology, incorporating: (1) semantic-driven entity and relation extraction optimized for medical terminology, (2) multi-dimensional decision relationship reconstruction from unstructured medical texts, and (3) human-guided reasoning for knowledge expansion. KG4Diagnosis serves as an extensible foundation for specialized medical diagnosis systems, with capabilities to incorporate new diseases and medical knowledge. The framework's modular design enables seamless integration of domain-specific enhancements, making it valuable for developing targeted medical diagnosis systems. We provide architectural guidelines and protocols to facilitate adoption across medical contexts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge