Junwei Yang
Kimi K2.5: Visual Agentic Intelligence
Feb 02, 2026Abstract:We introduce Kimi K2.5, an open-source multimodal agentic model designed to advance general agentic intelligence. K2.5 emphasizes the joint optimization of text and vision so that two modalities enhance each other. This includes a series of techniques such as joint text-vision pre-training, zero-vision SFT, and joint text-vision reinforcement learning. Building on this multimodal foundation, K2.5 introduces Agent Swarm, a self-directed parallel agent orchestration framework that dynamically decomposes complex tasks into heterogeneous sub-problems and executes them concurrently. Extensive evaluations show that Kimi K2.5 achieves state-of-the-art results across various domains including coding, vision, reasoning, and agentic tasks. Agent Swarm also reduces latency by up to $4.5\times$ over single-agent baselines. We release the post-trained Kimi K2.5 model checkpoint to facilitate future research and real-world applications of agentic intelligence.
ILLUME+: Illuminating Unified MLLM with Dual Visual Tokenization and Diffusion Refinement
Apr 03, 2025



Abstract:We present ILLUME+ that leverages dual visual tokenization and a diffusion decoder to improve both deep semantic understanding and high-fidelity image generation. Existing unified models have struggled to simultaneously handle the three fundamental capabilities in a unified model: understanding, generation, and editing. Models like Chameleon and EMU3 utilize VQGAN for image discretization, due to the lack of deep semantic interaction, they lag behind specialist models like LLaVA in visual understanding tasks. To mitigate this, LaViT and ILLUME employ semantic encoders for tokenization, but they struggle with image editing due to poor texture preservation. Meanwhile, Janus series decouples the input and output image representation, limiting their abilities to seamlessly handle interleaved image-text understanding and generation. In contrast, ILLUME+ introduces a unified dual visual tokenizer, DualViTok, which preserves both fine-grained textures and text-aligned semantics while enabling a coarse-to-fine image representation strategy for multimodal understanding and generation. Additionally, we employ a diffusion model as the image detokenizer for enhanced generation quality and efficient super-resolution. ILLUME+ follows a continuous-input, discrete-output scheme within the unified MLLM and adopts a progressive training procedure that supports dynamic resolution across the vision tokenizer, MLLM, and diffusion decoder. This design allows for flexible and efficient context-aware image editing and generation across diverse tasks. ILLUME+ (3B) exhibits competitive performance against existing unified MLLMs and specialized models across multimodal understanding, generation, and editing benchmarks. With its strong performance, ILLUME+ provides a scalable and versatile foundation for future multimodal applications. Project Page: https://illume-unified-mllm.github.io/.
Large Language Model Agent: A Survey on Methodology, Applications and Challenges
Mar 27, 2025Abstract:The era of intelligent agents is upon us, driven by revolutionary advancements in large language models. Large Language Model (LLM) agents, with goal-driven behaviors and dynamic adaptation capabilities, potentially represent a critical pathway toward artificial general intelligence. This survey systematically deconstructs LLM agent systems through a methodology-centered taxonomy, linking architectural foundations, collaboration mechanisms, and evolutionary pathways. We unify fragmented research threads by revealing fundamental connections between agent design principles and their emergent behaviors in complex environments. Our work provides a unified architectural perspective, examining how agents are constructed, how they collaborate, and how they evolve over time, while also addressing evaluation methodologies, tool applications, practical challenges, and diverse application domains. By surveying the latest developments in this rapidly evolving field, we offer researchers a structured taxonomy for understanding LLM agents and identify promising directions for future research. The collection is available at https://github.com/luo-junyu/Awesome-Agent-Papers.
ILLUME: Illuminating Your LLMs to See, Draw, and Self-Enhance
Dec 09, 2024Abstract:In this paper, we introduce ILLUME, a unified multimodal large language model (MLLM) that seamlessly integrates multimodal understanding and generation capabilities within a single large language model through a unified next-token prediction formulation. To address the large dataset size typically required for image-text alignment, we propose to enhance data efficiency through the design of a vision tokenizer that incorporates semantic information and a progressive multi-stage training procedure. This approach reduces the dataset size to just 15M for pretraining -- over four times fewer than what is typically needed -- while achieving competitive or even superior performance with existing unified MLLMs, such as Janus. Additionally, to promote synergistic enhancement between understanding and generation capabilities, which is under-explored in previous works, we introduce a novel self-enhancing multimodal alignment scheme. This scheme supervises the MLLM to self-assess the consistency between text descriptions and self-generated images, facilitating the model to interpret images more accurately and avoid unrealistic and incorrect predictions caused by misalignment in image generation. Based on extensive experiments, our proposed ILLUME stands out and competes with state-of-the-art unified MLLMs and specialized models across various benchmarks for multimodal understanding, generation, and editing.
SMI-Editor: Edit-based SMILES Language Model with Fragment-level Supervision
Dec 07, 2024



Abstract:SMILES, a crucial textual representation of molecular structures, has garnered significant attention as a foundation for pre-trained language models (LMs). However, most existing pre-trained SMILES LMs focus solely on the single-token level supervision during pre-training, failing to fully leverage the substructural information of molecules. This limitation makes the pre-training task overly simplistic, preventing the models from capturing richer molecular semantic information. Moreover, during pre-training, these SMILES LMs only process corrupted SMILES inputs, never encountering any valid SMILES, which leads to a train-inference mismatch. To address these challenges, we propose SMI-Editor, a novel edit-based pre-trained SMILES LM. SMI-Editor disrupts substructures within a molecule at random and feeds the resulting SMILES back into the model, which then attempts to restore the original SMILES through an editing process. This approach not only introduces fragment-level training signals, but also enables the use of valid SMILES as inputs, allowing the model to learn how to reconstruct complete molecules from these incomplete structures. As a result, the model demonstrates improved scalability and an enhanced ability to capture fragment-level molecular information. Experimental results show that SMI-Editor achieves state-of-the-art performance across multiple downstream molecular tasks, and even outperforming several 3D molecular representation models.
Towards Graph Contrastive Learning: A Survey and Beyond
May 20, 2024



Abstract:In recent years, deep learning on graphs has achieved remarkable success in various domains. However, the reliance on annotated graph data remains a significant bottleneck due to its prohibitive cost and time-intensive nature. To address this challenge, self-supervised learning (SSL) on graphs has gained increasing attention and has made significant progress. SSL enables machine learning models to produce informative representations from unlabeled graph data, reducing the reliance on expensive labeled data. While SSL on graphs has witnessed widespread adoption, one critical component, Graph Contrastive Learning (GCL), has not been thoroughly investigated in the existing literature. Thus, this survey aims to fill this gap by offering a dedicated survey on GCL. We provide a comprehensive overview of the fundamental principles of GCL, including data augmentation strategies, contrastive modes, and contrastive optimization objectives. Furthermore, we explore the extensions of GCL to other aspects of data-efficient graph learning, such as weakly supervised learning, transfer learning, and related scenarios. We also discuss practical applications spanning domains such as drug discovery, genomics analysis, recommender systems, and finally outline the challenges and potential future directions in this field.
Unsupervised Adaptive Implicit Neural Representation Learning for Scan-Specific MRI Reconstruction
Dec 01, 2023Abstract:In recent studies on MRI reconstruction, advances have shown significant promise for further accelerating the MRI acquisition. Most state-of-the-art methods require a large amount of fully-sampled data to optimise reconstruction models, which is impractical and expensive under certain clinical settings. On the other hand, for unsupervised scan-specific reconstruction methods, overfitting is likely to happen due to insufficient supervision, while restrictions on acceleration rates and under-sampling patterns further limit their applicability. To this end, we propose an unsupervised, adaptive coarse-to-fine framework that enhances reconstruction quality without being constrained by the sparsity levels or patterns in under-sampling. The framework employs an implicit neural representation for scan-specific MRI reconstruction, learning a mapping from multi-dimensional coordinates to their corresponding signal intensities. Moreover, we integrate a novel learning strategy that progressively refines the use of acquired k-space signals for self-supervision. This approach effectively adjusts the proportion of supervising signals from unevenly distributed information across different frequency bands, thus mitigating the issue of overfitting while improving the overall reconstruction. Comprehensive evaluation on a public dataset, including both 2D and 3D data, has shown that our method outperforms current state-of-the-art scan-specific MRI reconstruction techniques, for up to 8-fold under-sampling.
Dual-Domain Multi-Contrast MRI Reconstruction with Synthesis-based Fusion Network
Dec 01, 2023



Abstract:Purpose: To develop an efficient dual-domain reconstruction framework for multi-contrast MRI, with the focus on minimising cross-contrast misalignment in both the image and the frequency domains to enhance optimisation. Theory and Methods: Our proposed framework, based on deep learning, facilitates the optimisation for under-sampled target contrast using fully-sampled reference contrast that is quicker to acquire. The method consists of three key steps: 1) Learning to synthesise data resembling the target contrast from the reference contrast; 2) Registering the multi-contrast data to reduce inter-scan motion; and 3) Utilising the registered data for reconstructing the target contrast. These steps involve learning in both domains with regularisation applied to ensure their consistency. We also compare the reconstruction performance with existing deep learning-based methods using a dataset of brain MRI scans. Results: Extensive experiments demonstrate the superiority of our proposed framework, for up to an 8-fold acceleration rate, compared to state-of-the-art algorithms. Comprehensive analysis and ablation studies further present the effectiveness of the proposed components. Conclusion:Our dual-domain framework offers a promising approach to multi-contrast MRI reconstruction. It can also be integrated with existing methods to further enhance the reconstruction.
A Comprehensive Survey on Deep Graph Representation Learning
Apr 19, 2023



Abstract:Graph representation learning aims to effectively encode high-dimensional sparse graph-structured data into low-dimensional dense vectors, which is a fundamental task that has been widely studied in a range of fields, including machine learning and data mining. Classic graph embedding methods follow the basic idea that the embedding vectors of interconnected nodes in the graph can still maintain a relatively close distance, thereby preserving the structural information between the nodes in the graph. However, this is sub-optimal due to: (i) traditional methods have limited model capacity which limits the learning performance; (ii) existing techniques typically rely on unsupervised learning strategies and fail to couple with the latest learning paradigms; (iii) representation learning and downstream tasks are dependent on each other which should be jointly enhanced. With the remarkable success of deep learning, deep graph representation learning has shown great potential and advantages over shallow (traditional) methods, there exist a large number of deep graph representation learning techniques have been proposed in the past decade, especially graph neural networks. In this survey, we conduct a comprehensive survey on current deep graph representation learning algorithms by proposing a new taxonomy of existing state-of-the-art literature. Specifically, we systematically summarize the essential components of graph representation learning and categorize existing approaches by the ways of graph neural network architectures and the most recent advanced learning paradigms. Moreover, this survey also provides the practical and promising applications of deep graph representation learning. Last but not least, we state new perspectives and suggest challenging directions which deserve further investigations in the future.
MetaFill: Text Infilling for Meta-Path Generation on Heterogeneous Information Networks
Oct 14, 2022
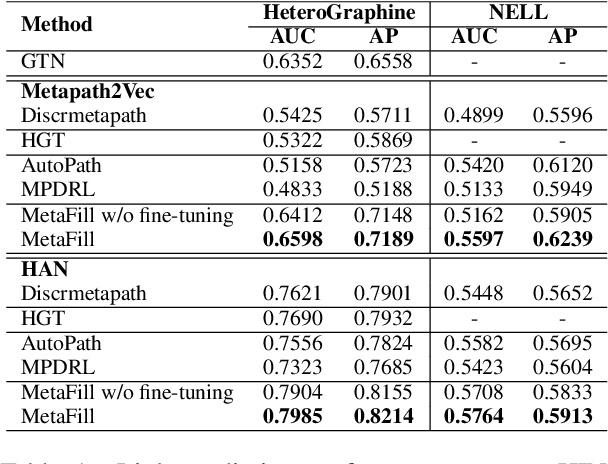


Abstract:Heterogeneous Information Network (HIN) is essential to study complicated networks containing multiple edge types and node types. Meta-path, a sequence of node types and edge types, is the core technique to embed HINs. Since manually curating meta-paths is time-consuming, there is a pressing need to develop automated meta-path generation approaches. Existing meta-path generation approaches cannot fully exploit the rich textual information in HINs, such as node names and edge type names. To address this problem, we propose MetaFill, a text-infilling-based approach for meta-path generation. The key idea of MetaFill is to formulate meta-path identification problem as a word sequence infilling problem, which can be advanced by Pretrained Language Models (PLMs). We observed the superior performance of MetaFill against existing meta-path generation methods and graph embedding methods that do not leverage meta-paths in both link prediction and node classification on two real-world HIN datasets. We further demonstrated how MetaFill can accurately classify edges in the zero-shot setting, where existing approaches cannot generate any meta-paths. MetaFill exploits PLMs to generate meta-paths for graph embedding, opening up new avenues for language model applications in graph analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge