Meng Qu
AdaCoT: Pareto-Optimal Adaptive Chain-of-Thought Triggering via Reinforcement Learning
May 17, 2025Abstract:Large Language Models (LLMs) have demonstrated remarkable capabilities but often face challenges with tasks requiring sophisticated reasoning. While Chain-of-Thought (CoT) prompting significantly enhances reasoning, it indiscriminately generates lengthy reasoning steps for all queries, leading to substantial computational costs and inefficiency, especially for simpler inputs. To address this critical issue, we introduce AdaCoT (Adaptive Chain-of-Thought), a novel framework enabling LLMs to adaptively decide when to invoke CoT. AdaCoT framed adaptive reasoning as a Pareto optimization problem that seeks to balance model performance with the costs associated with CoT invocation (both frequency and computational overhead). We propose a reinforcement learning (RL) based method, specifically utilizing Proximal Policy Optimization (PPO), to dynamically control the CoT triggering decision boundary by adjusting penalty coefficients, thereby allowing the model to determine CoT necessity based on implicit query complexity. A key technical contribution is Selective Loss Masking (SLM), designed to counteract decision boundary collapse during multi-stage RL training, ensuring robust and stable adaptive triggering. Experimental results demonstrate that AdaCoT successfully navigates the Pareto frontier, achieving substantial reductions in CoT usage for queries not requiring elaborate reasoning. For instance, on our production traffic testset, AdaCoT reduced CoT triggering rates to as low as 3.18\% and decreased average response tokens by 69.06%, while maintaining high performance on complex tasks.
Prioritizing Safeguarding Over Autonomy: Risks of LLM Agents for Science
Feb 07, 2024



Abstract:Intelligent agents powered by large language models (LLMs) have demonstrated substantial promise in autonomously conducting experiments and facilitating scientific discoveries across various disciplines. While their capabilities are promising, they also introduce novel vulnerabilities that demand careful consideration for safety. However, there exists a notable gap in the literature, as there has been no comprehensive exploration of these vulnerabilities. This position paper fills this gap by conducting a thorough examination of vulnerabilities in LLM-based agents within scientific domains, shedding light on potential risks associated with their misuse and emphasizing the need for safety measures. We begin by providing a comprehensive overview of the potential risks inherent to scientific LLM agents, taking into account user intent, the specific scientific domain, and their potential impact on the external environment. Then, we delve into the origins of these vulnerabilities and provide a scoping review of the limited existing works. Based on our analysis, we propose a triadic framework involving human regulation, agent alignment, and an understanding of environmental feedback (agent regulation) to mitigate these identified risks. Furthermore, we highlight the limitations and challenges associated with safeguarding scientific agents and advocate for the development of improved models, robust benchmarks, and comprehensive regulations to address these issues effectively.
GraphText: Graph Reasoning in Text Space
Oct 02, 2023



Abstract:Large Language Models (LLMs) have gained the ability to assimilate human knowledge and facilitate natural language interactions with both humans and other LLMs. However, despite their impressive achievements, LLMs have not made significant advancements in the realm of graph machine learning. This limitation arises because graphs encapsulate distinct relational data, making it challenging to transform them into natural language that LLMs understand. In this paper, we bridge this gap with a novel framework, GraphText, that translates graphs into natural language. GraphText derives a graph-syntax tree for each graph that encapsulates both the node attributes and inter-node relationships. Traversal of the tree yields a graph text sequence, which is then processed by an LLM to treat graph tasks as text generation tasks. Notably, GraphText offers multiple advantages. It introduces training-free graph reasoning: even without training on graph data, GraphText with ChatGPT can achieve on par with, or even surpassing, the performance of supervised-trained graph neural networks through in-context learning (ICL). Furthermore, GraphText paves the way for interactive graph reasoning, allowing both humans and LLMs to communicate with the model seamlessly using natural language. These capabilities underscore the vast, yet-to-be-explored potential of LLMs in the domain of graph machine learning.
TGNN: A Joint Semi-supervised Framework for Graph-level Classification
Apr 23, 2023Abstract:This paper studies semi-supervised graph classification, a crucial task with a wide range of applications in social network analysis and bioinformatics. Recent works typically adopt graph neural networks to learn graph-level representations for classification, failing to explicitly leverage features derived from graph topology (e.g., paths). Moreover, when labeled data is scarce, these methods are far from satisfactory due to their insufficient topology exploration of unlabeled data. We address the challenge by proposing a novel semi-supervised framework called Twin Graph Neural Network (TGNN). To explore graph structural information from complementary views, our TGNN has a message passing module and a graph kernel module. To fully utilize unlabeled data, for each module, we calculate the similarity of each unlabeled graph to other labeled graphs in the memory bank and our consistency loss encourages consistency between two similarity distributions in different embedding spaces. The two twin modules collaborate with each other by exchanging instance similarity knowledge to fully explore the structure information of both labeled and unlabeled data. We evaluate our TGNN on various public datasets and show that it achieves strong performance.
Learning on Large-scale Text-attributed Graphs via Variational Inference
Oct 26, 2022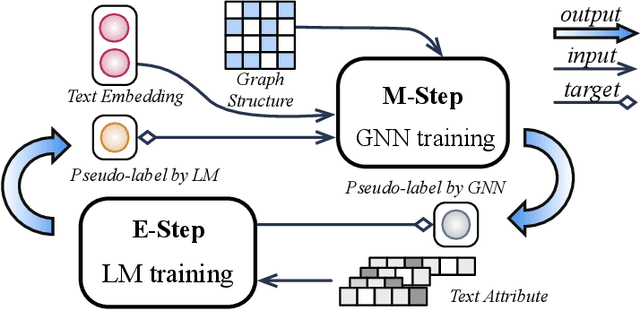
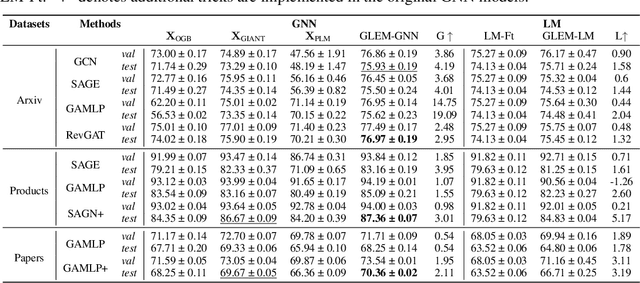
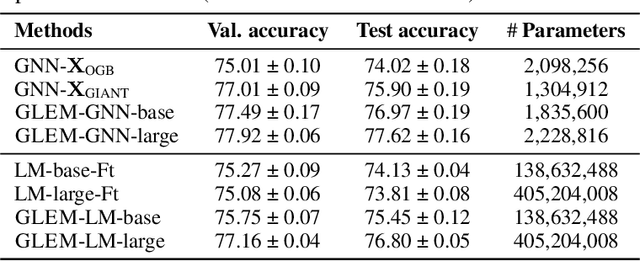
Abstract:This paper studies learning on text-attributed graphs (TAGs), where each node is associated with a text description. An ideal solution for such a problem would be integrating both the text and graph structure information with large language models and graph neural networks (GNNs). However, the problem becomes very challenging when graphs are large due to the high computational complexity brought by large language models and training GNNs on big graphs. In this paper, we propose an efficient and effective solution to learning on large text-attributed graphs by fusing graph structure and language learning with a variational Expectation-Maximization (EM) framework, called GLEM. Instead of simultaneously training large language models and GNNs on big graphs, GLEM proposes to alternatively update the two modules in the E-step and M-step. Such a procedure allows to separately train the two modules but at the same time allows the two modules to interact and mutually enhance each other. Extensive experiments on multiple data sets demonstrate the efficiency and effectiveness of the proposed approach.
KGNN: Harnessing Kernel-based Networks for Semi-supervised Graph Classification
May 21, 2022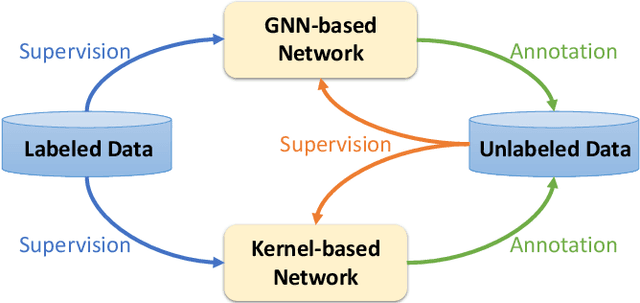
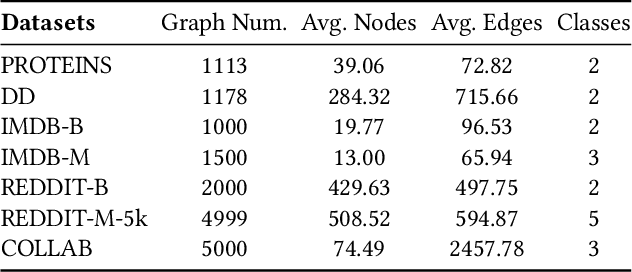
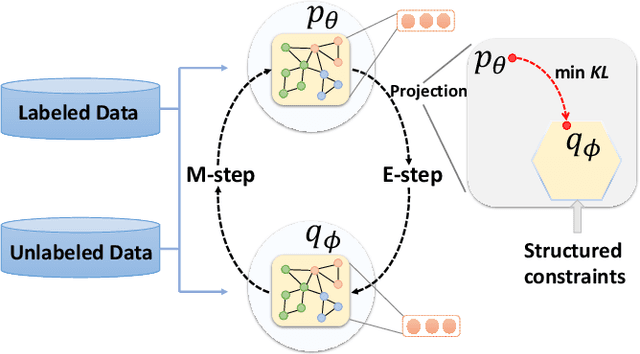
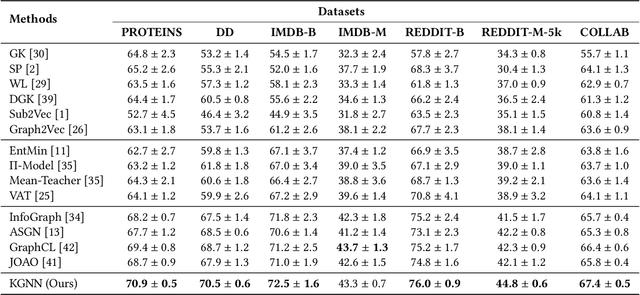
Abstract:This paper studies semi-supervised graph classification, which is an important problem with various applications in social network analysis and bioinformatics. This problem is typically solved by using graph neural networks (GNNs), which yet rely on a large number of labeled graphs for training and are unable to leverage unlabeled graphs. We address the limitations by proposing the Kernel-based Graph Neural Network (KGNN). A KGNN consists of a GNN-based network as well as a kernel-based network parameterized by a memory network. The GNN-based network performs classification through learning graph representations to implicitly capture the similarity between query graphs and labeled graphs, while the kernel-based network uses graph kernels to explicitly compare each query graph with all the labeled graphs stored in a memory for prediction. The two networks are motivated from complementary perspectives, and thus combing them allows KGNN to use labeled graphs more effectively. We jointly train the two networks by maximizing their agreement on unlabeled graphs via posterior regularization, so that the unlabeled graphs serve as a bridge to let both networks mutually enhance each other. Experiments on a range of well-known benchmark datasets demonstrate that KGNN achieves impressive performance over competitive baselines.
Neural Structured Prediction for Inductive Node Classification
Apr 15, 2022



Abstract:This paper studies node classification in the inductive setting, i.e., aiming to learn a model on labeled training graphs and generalize it to infer node labels on unlabeled test graphs. This problem has been extensively studied with graph neural networks (GNNs) by learning effective node representations, as well as traditional structured prediction methods for modeling the structured output of node labels, e.g., conditional random fields (CRFs). In this paper, we present a new approach called the Structured Proxy Network (SPN), which combines the advantages of both worlds. SPN defines flexible potential functions of CRFs with GNNs. However, learning such a model is nontrivial as it involves optimizing a maximin game with high-cost inference. Inspired by the underlying connection between joint and marginal distributions defined by Markov networks, we propose to solve an approximate version of the optimization problem as a proxy, which yields a near-optimal solution, making learning more efficient. Extensive experiments on two settings show that our approach outperforms many competitive baselines.
Structured Multi-task Learning for Molecular Property Prediction
Feb 22, 2022



Abstract:Multi-task learning for molecular property prediction is becoming increasingly important in drug discovery. However, in contrast to other domains, the performance of multi-task learning in drug discovery is still not satisfying as the number of labeled data for each task is too limited, which calls for additional data to complement the data scarcity. In this paper, we study multi-task learning for molecular property prediction in a novel setting, where a relation graph between tasks is available. We first construct a dataset including around 400 tasks as well as a task relation graph. Then to better utilize such relation graph, we propose a method called SGNN-EBM to systematically investigate the structured task modeling from two perspectives. (1) In the \emph{latent} space, we model the task representations by applying a state graph neural network (SGNN) on the relation graph. (2) In the \emph{output} space, we employ structured prediction with the energy-based model (EBM), which can be efficiently trained through noise-contrastive estimation (NCE) approach. Empirical results justify the effectiveness of SGNN-EBM. Code is available on https://github.com/chao1224/SGNN-EBM.
TorchDrug: A Powerful and Flexible Machine Learning Platform for Drug Discovery
Feb 16, 2022

Abstract:Machine learning has huge potential to revolutionize the field of drug discovery and is attracting increasing attention in recent years. However, lacking domain knowledge (e.g., which tasks to work on), standard benchmarks and data preprocessing pipelines are the main obstacles for machine learning researchers to work in this domain. To facilitate the progress of machine learning for drug discovery, we develop TorchDrug, a powerful and flexible machine learning platform for drug discovery built on top of PyTorch. TorchDrug benchmarks a variety of important tasks in drug discovery, including molecular property prediction, pretrained molecular representations, de novo molecular design and optimization, retrosynthsis prediction, and biomedical knowledge graph reasoning. State-of-the-art techniques based on geometric deep learning (or graph machine learning), deep generative models, reinforcement learning and knowledge graph reasoning are implemented for these tasks. TorchDrug features a hierarchical interface that facilitates customization from both novices and experts in this domain. Tutorials, benchmark results and documentation are available at https://torchdrug.ai. Code is released under Apache License 2.0.
COVI-AgentSim: an Agent-based Model for Evaluating Methods of Digital Contact Tracing
Oct 30, 2020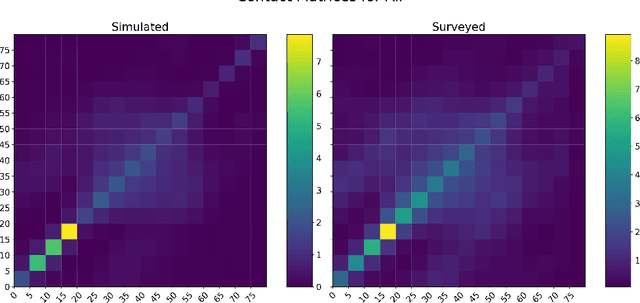
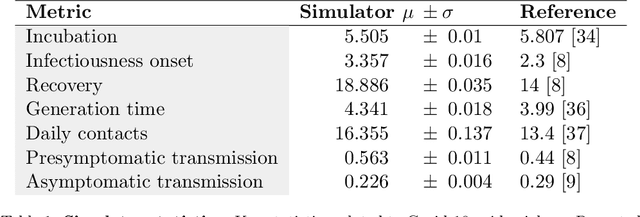
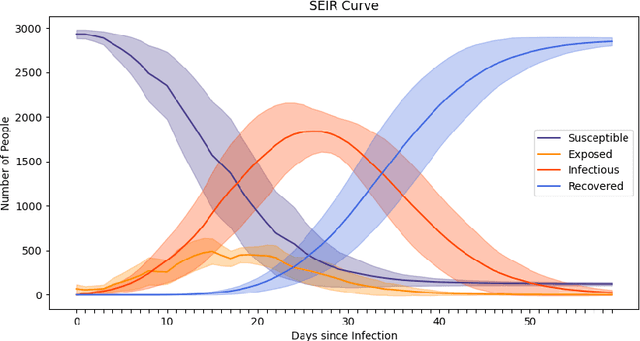
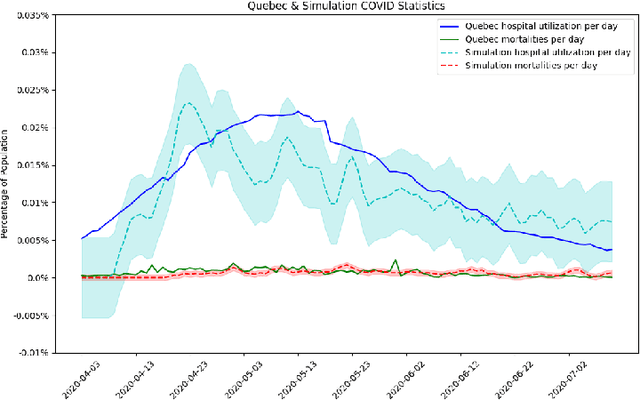
Abstract:The rapid global spread of COVID-19 has led to an unprecedented demand for effective methods to mitigate the spread of the disease, and various digital contact tracing (DCT) methods have emerged as a component of the solution. In order to make informed public health choices, there is a need for tools which allow evaluation and comparison of DCT methods. We introduce an agent-based compartmental simulator we call COVI-AgentSim, integrating detailed consideration of virology, disease progression, social contact networks, and mobility patterns, based on parameters derived from empirical research. We verify by comparing to real data that COVI-AgentSim is able to reproduce realistic COVID-19 spread dynamics, and perform a sensitivity analysis to verify that the relative performance of contact tracing methods are consistent across a range of settings. We use COVI-AgentSim to perform cost-benefit analyses comparing no DCT to: 1) standard binary contact tracing (BCT) that assigns binary recommendations based on binary test results; and 2) a rule-based method for feature-based contact tracing (FCT) that assigns a graded level of recommendation based on diverse individual features. We find all DCT methods consistently reduce the spread of the disease, and that the advantage of FCT over BCT is maintained over a wide range of adoption rates. Feature-based methods of contact tracing avert more disability-adjusted life years (DALYs) per socioeconomic cost (measured by productive hours lost). Our results suggest any DCT method can help save lives, support re-opening of economies, and prevent second-wave outbreaks, and that FCT methods are a promising direction for enriching BCT using self-reported symptoms, yielding earlier warning signals and a significantly reduced spread of the virus per socioeconomic cost.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge