Abhinav Sharma
The Llama 4 Herd: Architecture, Training, Evaluation, and Deployment Notes
Jan 15, 2026Abstract:This document consolidates publicly reported technical details about Metas Llama 4 model family. It summarizes (i) released variants (Scout and Maverick) and the broader herd context including the previewed Behemoth teacher model, (ii) architectural characteristics beyond a high-level MoE description covering routed/shared-expert structure, early-fusion multimodality, and long-context design elements reported for Scout (iRoPE and length generalization strategies), (iii) training disclosures spanning pre-training, mid-training for long-context extension, and post-training methodology (lightweight SFT, online RL, and lightweight DPO) as described in release materials, (iv) developer-reported benchmark results for both base and instruction-tuned checkpoints, and (v) practical deployment constraints observed across major serving environments, including provider-specific context limits and quantization packaging. The manuscript also summarizes licensing obligations relevant to redistribution and derivative naming, and reviews publicly described safeguards and evaluation practices. The goal is to provide a compact technical reference for researchers and practitioners who need precise, source-backed facts about Llama 4.
ASSERTIFY: Utilizing Large Language Models to Generate Assertions for Production Code
Nov 25, 2024Abstract:Production assertions are statements embedded in the code to help developers validate their assumptions about the code. They assist developers in debugging, provide valuable documentation, and enhance code comprehension. Current research in this area primarily focuses on assertion generation for unit tests using techniques, such as static analysis and deep learning. While these techniques have shown promise, they fall short when it comes to generating production assertions, which serve a different purpose. This preprint addresses the gap by introducing Assertify, an automated end-to-end tool that leverages Large Language Models (LLMs) and prompt engineering with few-shot learning to generate production assertions. By creating context-rich prompts, the tool emulates the approach developers take when creating production assertions for their code. To evaluate our approach, we compiled a dataset of 2,810 methods by scraping 22 mature Java repositories from GitHub. Our experiments demonstrate the effectiveness of few-shot learning by producing assertions with an average ROUGE-L score of 0.526, indicating reasonably high structural similarity with the assertions written by developers. This research demonstrates the potential of LLMs in automating the generation of production assertions that resemble the original assertions.
Representation Similarity: A Better Guidance of DNN Layer Sharing for Edge Computing without Training
Oct 15, 2024Abstract:Edge computing has emerged as an alternative to reduce transmission and processing delay and preserve privacy of the video streams. However, the ever-increasing complexity of Deep Neural Networks (DNNs) used in video-based applications (e.g. object detection) exerts pressure on memory-constrained edge devices. Model merging is proposed to reduce the DNNs' memory footprint by keeping only one copy of merged layers' weights in memory. In existing model merging techniques, (i) only architecturally identical layers can be shared; (ii) requires computationally expensive retraining in the cloud; (iii) assumes the availability of ground truth for retraining. The re-evaluation of a merged model's performance, however, requires a validation dataset with ground truth, typically runs at the cloud. Common metrics to guide the selection of shared layers include the size or computational cost of shared layers or representation size. We propose a new model merging scheme by sharing representations (i.e., outputs of layers) at the edge, guided by representation similarity S. We show that S is extremely highly correlated with merged model's accuracy with Pearson Correlation Coefficient |r| > 0.94 than other metrics, demonstrating that representation similarity can serve as a strong validation accuracy indicator without ground truth. We present our preliminary results of the newly proposed model merging scheme with identified challenges, demonstrating a promising research future direction.
A Lightweight Measure of Classification Difficulty from Application Dataset Characteristics
Apr 09, 2024



Abstract:Despite accuracy and computation benchmarks being widely available to help choose among neural network models, these are usually trained on datasets with many classes, and do not give a precise idea of performance for applications of few (< 10) classes. The conventional procedure to predict performance is to train and test repeatedly on the different models and dataset variations of interest. However, this is computationally expensive. We propose an efficient classification difficulty measure that is calculated from the number of classes and intra- and inter-class similarity metrics of the dataset. After a single stage of training and testing per model family, relative performance for different datasets and models of the same family can be predicted by comparing difficulty measures - without further training and testing. We show how this measure can help a practitioner select a computationally efficient model for a small dataset 6 to 29x faster than through repeated training and testing. We give an example of use of the measure for an industrial application in which options are identified to select a model 42% smaller than the baseline YOLOv5-nano model, and if class merging from 3 to 2 classes meets requirements, 85% smaller.
WEEP: A method for spatial interpretation of weakly supervised CNN models in computational pathology
Apr 08, 2024Abstract:Deep learning enables the modelling of high-resolution histopathology whole-slide images (WSI). Weakly supervised learning of tile-level data is typically applied for tasks where labels only exist on the patient or WSI level (e.g. patient outcomes or histological grading). In this context, there is a need for improved spatial interpretability of predictions from such models. We propose a novel method, Wsi rEgion sElection aPproach (WEEP), for model interpretation. It provides a principled yet straightforward way to establish the spatial area of WSI required for assigning a particular prediction label. We demonstrate WEEP on a binary classification task in the area of breast cancer computational pathology. WEEP is easy to implement, is directly connected to the model-based decision process, and offers information relevant to both research and diagnostic applications.
The ACROBAT 2022 Challenge: Automatic Registration Of Breast Cancer Tissue
May 29, 2023Abstract:The alignment of tissue between histopathological whole-slide-images (WSI) is crucial for research and clinical applications. Advances in computing, deep learning, and availability of large WSI datasets have revolutionised WSI analysis. Therefore, the current state-of-the-art in WSI registration is unclear. To address this, we conducted the ACROBAT challenge, based on the largest WSI registration dataset to date, including 4,212 WSIs from 1,152 breast cancer patients. The challenge objective was to align WSIs of tissue that was stained with routine diagnostic immunohistochemistry to its H&E-stained counterpart. We compare the performance of eight WSI registration algorithms, including an investigation of the impact of different WSI properties and clinical covariates. We find that conceptually distinct WSI registration methods can lead to highly accurate registration performances and identify covariates that impact performances across methods. These results establish the current state-of-the-art in WSI registration and guide researchers in selecting and developing methods.
ACROBAT -- a multi-stain breast cancer histological whole-slide-image data set from routine diagnostics for computational pathology
Nov 24, 2022Abstract:The analysis of FFPE tissue sections stained with haematoxylin and eosin (H&E) or immunohistochemistry (IHC) is an essential part of the pathologic assessment of surgically resected breast cancer specimens. IHC staining has been broadly adopted into diagnostic guidelines and routine workflows to manually assess status and scoring of several established biomarkers, including ER, PGR, HER2 and KI67. However, this is a task that can also be facilitated by computational pathology image analysis methods. The research in computational pathology has recently made numerous substantial advances, often based on publicly available whole slide image (WSI) data sets. However, the field is still considerably limited by the sparsity of public data sets. In particular, there are no large, high quality publicly available data sets with WSIs of matching IHC and H&E-stained tissue sections. Here, we publish the currently largest publicly available data set of WSIs of tissue sections from surgical resection specimens from female primary breast cancer patients with matched WSIs of corresponding H&E and IHC-stained tissue, consisting of 4,212 WSIs from 1,153 patients. The primary purpose of the data set was to facilitate the ACROBAT WSI registration challenge, aiming at accurately aligning H&E and IHC images. For research in the area of image registration, automatic quantitative feedback on registration algorithm performance remains available through the ACROBAT challenge website, based on more than 37,000 manually annotated landmark pairs from 13 annotators. Beyond registration, this data set has the potential to enable many different avenues of computational pathology research, including stain-guided learning, virtual staining, unsupervised pre-training, artefact detection and stain-independent models.
Searching k-Optimal Goals for an Orienteering Problem on a Specialized Graph with Budget Constraints
Nov 02, 2020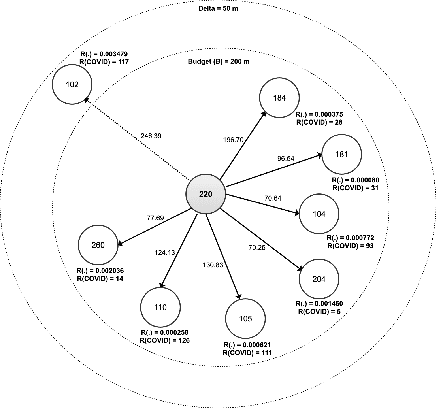
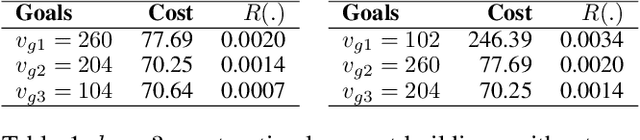

Abstract:We propose a novel non-randomized anytime orienteering algorithm for finding k-optimal goals that maximize reward on a specialized graph with budget constraints. This specialized graph represents a real-world scenario which is analogous to an orienteering problem of finding k-most optimal goal states.
COVI-AgentSim: an Agent-based Model for Evaluating Methods of Digital Contact Tracing
Oct 30, 2020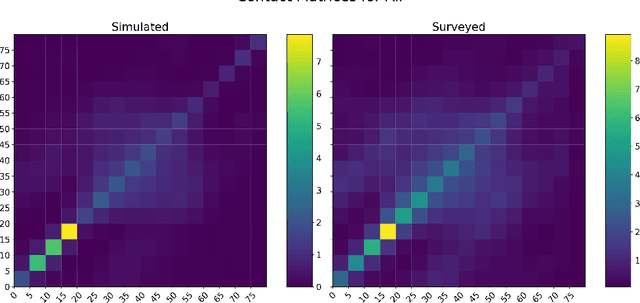
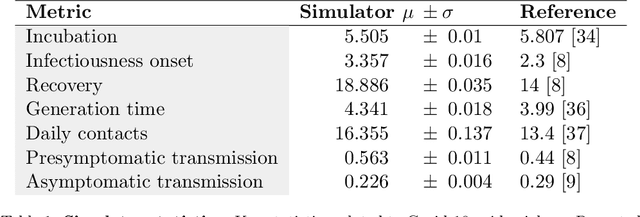
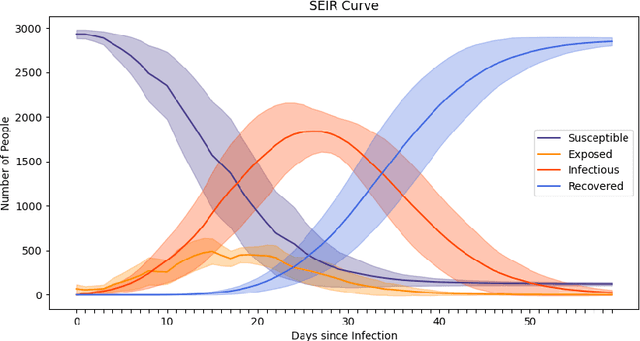
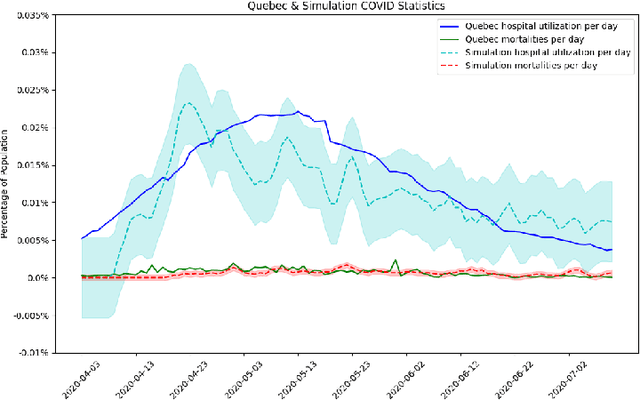
Abstract:The rapid global spread of COVID-19 has led to an unprecedented demand for effective methods to mitigate the spread of the disease, and various digital contact tracing (DCT) methods have emerged as a component of the solution. In order to make informed public health choices, there is a need for tools which allow evaluation and comparison of DCT methods. We introduce an agent-based compartmental simulator we call COVI-AgentSim, integrating detailed consideration of virology, disease progression, social contact networks, and mobility patterns, based on parameters derived from empirical research. We verify by comparing to real data that COVI-AgentSim is able to reproduce realistic COVID-19 spread dynamics, and perform a sensitivity analysis to verify that the relative performance of contact tracing methods are consistent across a range of settings. We use COVI-AgentSim to perform cost-benefit analyses comparing no DCT to: 1) standard binary contact tracing (BCT) that assigns binary recommendations based on binary test results; and 2) a rule-based method for feature-based contact tracing (FCT) that assigns a graded level of recommendation based on diverse individual features. We find all DCT methods consistently reduce the spread of the disease, and that the advantage of FCT over BCT is maintained over a wide range of adoption rates. Feature-based methods of contact tracing avert more disability-adjusted life years (DALYs) per socioeconomic cost (measured by productive hours lost). Our results suggest any DCT method can help save lives, support re-opening of economies, and prevent second-wave outbreaks, and that FCT methods are a promising direction for enriching BCT using self-reported symptoms, yielding earlier warning signals and a significantly reduced spread of the virus per socioeconomic cost.
Predicting Infectiousness for Proactive Contact Tracing
Oct 23, 2020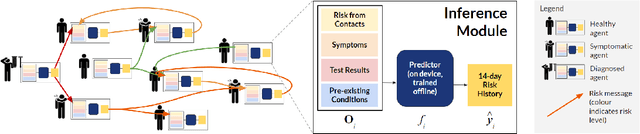
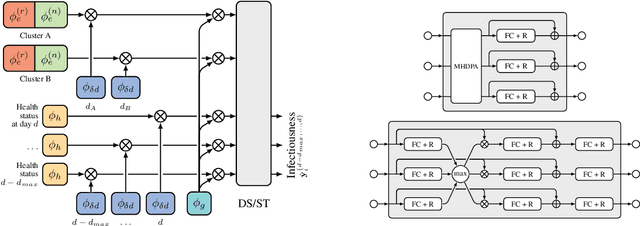
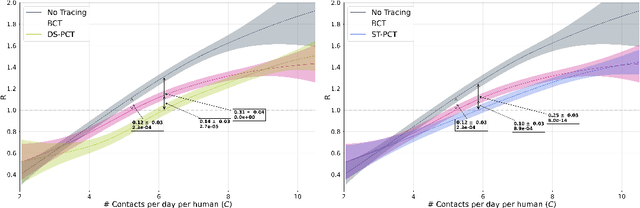
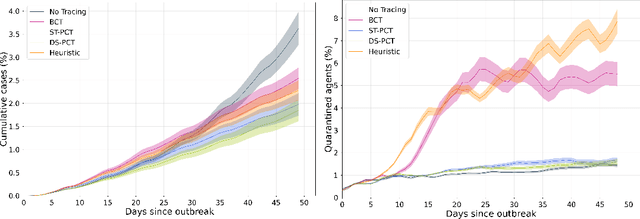
Abstract:The COVID-19 pandemic has spread rapidly worldwide, overwhelming manual contact tracing in many countries and resulting in widespread lockdowns for emergency containment. Large-scale digital contact tracing (DCT) has emerged as a potential solution to resume economic and social activity while minimizing spread of the virus. Various DCT methods have been proposed, each making trade-offs between privacy, mobility restrictions, and public health. The most common approach, binary contact tracing (BCT), models infection as a binary event, informed only by an individual's test results, with corresponding binary recommendations that either all or none of the individual's contacts quarantine. BCT ignores the inherent uncertainty in contacts and the infection process, which could be used to tailor messaging to high-risk individuals, and prompt proactive testing or earlier warnings. It also does not make use of observations such as symptoms or pre-existing medical conditions, which could be used to make more accurate infectiousness predictions. In this paper, we use a recently-proposed COVID-19 epidemiological simulator to develop and test methods that can be deployed to a smartphone to locally and proactively predict an individual's infectiousness (risk of infecting others) based on their contact history and other information, while respecting strong privacy constraints. Predictions are used to provide personalized recommendations to the individual via an app, as well as to send anonymized messages to the individual's contacts, who use this information to better predict their own infectiousness, an approach we call proactive contact tracing (PCT). We find a deep-learning based PCT method which improves over BCT for equivalent average mobility, suggesting PCT could help in safe re-opening and second-wave prevention.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge