Minghao Xu
HermesFlow: Seamlessly Closing the Gap in Multimodal Understanding and Generation
Feb 17, 2025Abstract:The remarkable success of the autoregressive paradigm has made significant advancement in Multimodal Large Language Models (MLLMs), with powerful models like Show-o, Transfusion and Emu3 achieving notable progress in unified image understanding and generation. For the first time, we uncover a common phenomenon: the understanding capabilities of MLLMs are typically stronger than their generative capabilities, with a significant gap between the two. Building on this insight, we propose HermesFlow, a simple yet general framework designed to seamlessly bridge the gap between understanding and generation in MLLMs. Specifically, we take the homologous data as input to curate homologous preference data of both understanding and generation. Through Pair-DPO and self-play iterative optimization, HermesFlow effectively aligns multimodal understanding and generation using homologous preference data. Extensive experiments demonstrate the significant superiority of our approach over prior methods, particularly in narrowing the gap between multimodal understanding and generation. These findings highlight the potential of HermesFlow as a general alignment framework for next-generation multimodal foundation models. Code: https://github.com/Gen-Verse/HermesFlow
No More Adam: Learning Rate Scaling at Initialization is All You Need
Dec 17, 2024Abstract:In this work, we question the necessity of adaptive gradient methods for training deep neural networks. SGD-SaI is a simple yet effective enhancement to stochastic gradient descent with momentum (SGDM). SGD-SaI performs learning rate Scaling at Initialization (SaI) to distinct parameter groups, guided by their respective gradient signal-to-noise ratios (g-SNR). By adjusting learning rates without relying on adaptive second-order momentum, SGD-SaI helps prevent training imbalances from the very first iteration and cuts the optimizer's memory usage by half compared to AdamW. Despite its simplicity and efficiency, SGD-SaI consistently matches or outperforms AdamW in training a variety of Transformer-based tasks, effectively overcoming a long-standing challenge of using SGD for training Transformers. SGD-SaI excels in ImageNet-1K classification with Vision Transformers(ViT) and GPT-2 pretraining for large language models (LLMs, transformer decoder-only), demonstrating robustness to hyperparameter variations and practicality for diverse applications. We further tested its robustness on tasks like LoRA fine-tuning for LLMs and diffusion models, where it consistently outperforms state-of-the-art optimizers. From a memory efficiency perspective, SGD-SaI achieves substantial memory savings for optimizer states, reducing memory usage by 5.93 GB for GPT-2 (1.5B parameters) and 25.15 GB for Llama2-7B compared to AdamW in full-precision training settings.
GlycanML: A Multi-Task and Multi-Structure Benchmark for Glycan Machine Learning
May 25, 2024



Abstract:Glycans are basic biomolecules and perform essential functions within living organisms. The rapid increase of functional glycan data provides a good opportunity for machine learning solutions to glycan understanding. However, there still lacks a standard machine learning benchmark for glycan function prediction. In this work, we fill this blank by building a comprehensive benchmark for Glycan Machine Learning (GlycanML). The GlycanML benchmark consists of diverse types of tasks including glycan taxonomy prediction, glycan immunogenicity prediction, glycosylation type prediction, and protein-glycan interaction prediction. Glycans can be represented by both sequences and graphs in GlycanML, which enables us to extensively evaluate sequence-based models and graph neural networks (GNNs) on benchmark tasks. Furthermore, by concurrently performing eight glycan taxonomy prediction tasks, we introduce the GlycanML-MTL testbed for multi-task learning (MTL) algorithms. Experimental results show the superiority of modeling glycans with multi-relational GNNs, and suitable MTL methods can further boost model performance. We provide all datasets and source codes at https://github.com/GlycanML/GlycanML and maintain a leaderboard at https://GlycanML.github.io/project
EditWorld: Simulating World Dynamics for Instruction-Following Image Editing
May 23, 2024



Abstract:Diffusion models have significantly improved the performance of image editing. Existing methods realize various approaches to achieve high-quality image editing, including but not limited to text control, dragging operation, and mask-and-inpainting. Among these, instruction-based editing stands out for its convenience and effectiveness in following human instructions across diverse scenarios. However, it still focuses on simple editing operations like adding, replacing, or deleting, and falls short of understanding aspects of world dynamics that convey the realistic dynamic nature in the physical world. Therefore, this work, EditWorld, introduces a new editing task, namely world-instructed image editing, which defines and categorizes the instructions grounded by various world scenarios. We curate a new image editing dataset with world instructions using a set of large pretrained models (e.g., GPT-3.5, Video-LLava and SDXL). To enable sufficient simulation of world dynamics for image editing, our EditWorld trains model in the curated dataset, and improves instruction-following ability with designed post-edit strategy. Extensive experiments demonstrate our method significantly outperforms existing editing methods in this new task. Our dataset and code will be available at https://github.com/YangLing0818/EditWorld
ProtLLM: An Interleaved Protein-Language LLM with Protein-as-Word Pre-Training
Feb 28, 2024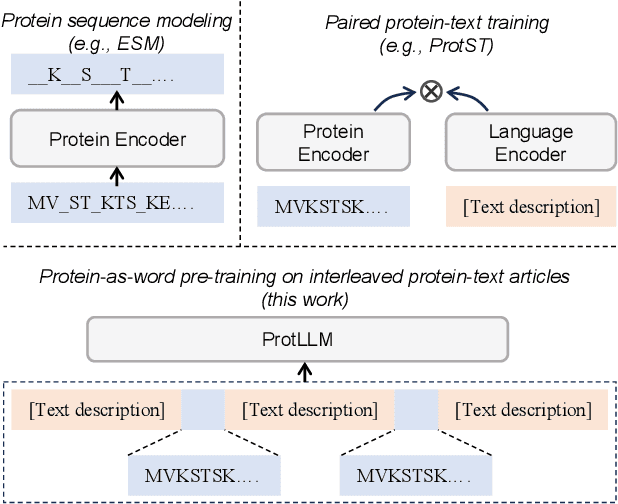

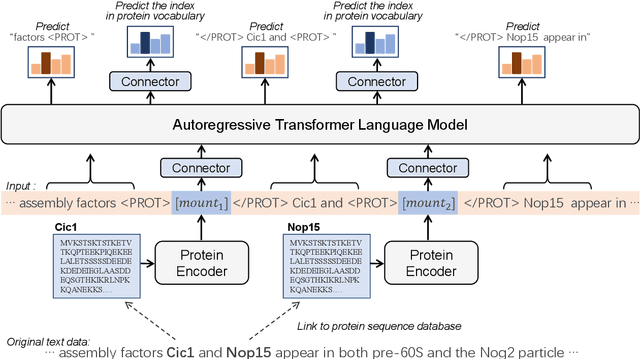

Abstract:We propose ProtLLM, a versatile cross-modal large language model (LLM) for both protein-centric and protein-language tasks. ProtLLM features a unique dynamic protein mounting mechanism, enabling it to handle complex inputs where the natural language text is interspersed with an arbitrary number of proteins. Besides, we propose the protein-as-word language modeling approach to train ProtLLM. By developing a specialized protein vocabulary, we equip the model with the capability to predict not just natural language but also proteins from a vast pool of candidates. Additionally, we construct a large-scale interleaved protein-text dataset, named InterPT, for pre-training. This dataset comprehensively encompasses both (1) structured data sources like protein annotations and (2) unstructured data sources like biological research papers, thereby endowing ProtLLM with crucial knowledge for understanding proteins. We evaluate ProtLLM on classic supervised protein-centric tasks and explore its novel protein-language applications. Experimental results demonstrate that ProtLLM not only achieves superior performance against protein-specialized baselines on protein-centric tasks but also induces zero-shot and in-context learning capabilities on protein-language tasks.
Enhancing Protein Language Models with Structure-based Encoder and Pre-training
Mar 11, 2023Abstract:Protein language models (PLMs) pre-trained on large-scale protein sequence corpora have achieved impressive performance on various downstream protein understanding tasks. Despite the ability to implicitly capture inter-residue contact information, transformer-based PLMs cannot encode protein structures explicitly for better structure-aware protein representations. Besides, the power of pre-training on available protein structures has not been explored for improving these PLMs, though structures are important to determine functions. To tackle these limitations, in this work, we enhance the PLMs with structure-based encoder and pre-training. We first explore feasible model architectures to combine the advantages of a state-of-the-art PLM (i.e., ESM-1b1) and a state-of-the-art protein structure encoder (i.e., GearNet). We empirically verify the ESM-GearNet that connects two encoders in a series way as the most effective combination model. To further improve the effectiveness of ESM-GearNet, we pre-train it on massive unlabeled protein structures with contrastive learning, which aligns representations of co-occurring subsequences so as to capture their biological correlation. Extensive experiments on EC and GO protein function prediction benchmarks demonstrate the superiority of ESM-GearNet over previous PLMs and structure encoders, and clear performance gains are further achieved by structure-based pre-training upon ESM-GearNet. Our implementation is available at https://github.com/DeepGraphLearning/GearNet.
Physics-Inspired Protein Encoder Pre-Training via Siamese Sequence-Structure Diffusion Trajectory Prediction
Jan 28, 2023Abstract:Pre-training methods on proteins are recently gaining interest, leveraging either protein sequences or structures, while modeling their joint energy landscape is largely unexplored. In this work, inspired by the success of denoising diffusion models, we propose the DiffPreT approach to pre-train a protein encoder by sequence-structure multimodal diffusion modeling. DiffPreT guides the encoder to recover the native protein sequences and structures from the perturbed ones along the multimodal diffusion trajectory, which acquires the joint distribution of sequences and structures. Considering the essential protein conformational variations, we enhance DiffPreT by a physics-inspired method called Siamese Diffusion Trajectory Prediction (SiamDiff) to capture the correlation between different conformers of a protein. SiamDiff attains this goal by maximizing the mutual information between representations of diffusion trajectories of structurally-correlated conformers. We study the effectiveness of DiffPreT and SiamDiff on both atom- and residue-level structure-based protein understanding tasks. Experimental results show that the performance of DiffPreT is consistently competitive on all tasks, and SiamDiff achieves new state-of-the-art performance, considering the mean ranks on all tasks. The source code will be released upon acceptance.
ProtST: Multi-Modality Learning of Protein Sequences and Biomedical Texts
Jan 28, 2023



Abstract:Current protein language models (PLMs) learn protein representations mainly based on their sequences, thereby well capturing co-evolutionary information, but they are unable to explicitly acquire protein functions, which is the end goal of protein representation learning. Fortunately, for many proteins, their textual property descriptions are available, where their various functions are also described. Motivated by this fact, we first build the ProtDescribe dataset to augment protein sequences with text descriptions of their functions and other important properties. Based on this dataset, we propose the ProtST framework to enhance Protein Sequence pre-training and understanding by biomedical Texts. During pre-training, we design three types of tasks, i.e., unimodal mask prediction, multimodal representation alignment and multimodal mask prediction, to enhance a PLM with protein property information with different granularities and, at the same time, preserve the PLM's original representation power. On downstream tasks, ProtST enables both supervised learning and zero-shot prediction. We verify the superiority of ProtST-induced PLMs over previous ones on diverse representation learning benchmarks. Under the zero-shot setting, we show the effectiveness of ProtST on zero-shot protein classification, and ProtST also enables functional protein retrieval from a large-scale database without any function annotation.
EurNet: Efficient Multi-Range Relational Modeling of Spatial Multi-Relational Data
Nov 23, 2022Abstract:Modeling spatial relationship in the data remains critical across many different tasks, such as image classification, semantic segmentation and protein structure understanding. Previous works often use a unified solution like relative positional encoding. However, there exists different kinds of spatial relations, including short-range, medium-range and long-range relations, and modeling them separately can better capture the focus of different tasks on the multi-range relations (e.g., short-range relations can be important in instance segmentation, while long-range relations should be upweighted for semantic segmentation). In this work, we introduce the EurNet for Efficient multi-range relational modeling. EurNet constructs the multi-relational graph, where each type of edge corresponds to short-, medium- or long-range spatial interactions. In the constructed graph, EurNet adopts a novel modeling layer, called gated relational message passing (GRMP), to propagate multi-relational information across the data. GRMP captures multiple relations within the data with little extra computational cost. We study EurNets in two important domains for image and protein structure modeling. Extensive experiments on ImageNet classification, COCO object detection and ADE20K semantic segmentation verify the gains of EurNet over the previous SoTA FocalNet. On the EC and GO protein function prediction benchmarks, EurNet consistently surpasses the previous SoTA GearNet. Our results demonstrate the strength of EurNets on modeling spatial multi-relational data from various domains. The implementations of EurNet for image modeling are available at https://github.com/hirl-team/EurNet-Image . The implementations for other applied domains/tasks will be released soon.
PEER: A Comprehensive and Multi-Task Benchmark for Protein Sequence Understanding
Jun 05, 2022


Abstract:We are now witnessing significant progress of deep learning methods in a variety of tasks (or datasets) of proteins. However, there is a lack of a standard benchmark to evaluate the performance of different methods, which hinders the progress of deep learning in this field. In this paper, we propose such a benchmark called PEER, a comprehensive and multi-task benchmark for Protein sEquence undERstanding. PEER provides a set of diverse protein understanding tasks including protein function prediction, protein localization prediction, protein structure prediction, protein-protein interaction prediction, and protein-ligand interaction prediction. We evaluate different types of sequence-based methods for each task including traditional feature engineering approaches, different sequence encoding methods as well as large-scale pre-trained protein language models. In addition, we also investigate the performance of these methods under the multi-task learning setting. Experimental results show that large-scale pre-trained protein language models achieve the best performance for most individual tasks, and jointly training multiple tasks further boosts the performance. The datasets and source codes of this benchmark will be open-sourced soon.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge