Payel Das
Patching LLM Like Software: A Lightweight Method for Improving Safety Policy in Large Language Models
Nov 11, 2025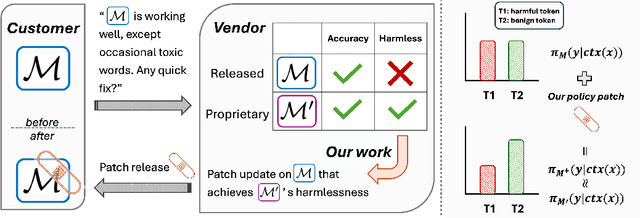
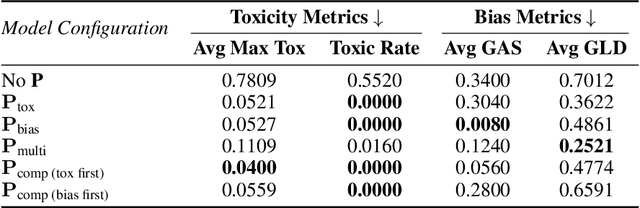
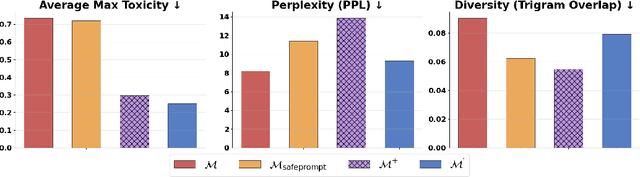
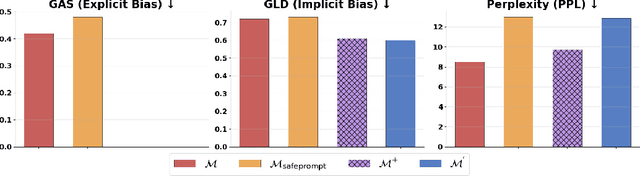
Abstract:We propose patching for large language models (LLMs) like software versions, a lightweight and modular approach for addressing safety vulnerabilities. While vendors release improved LLM versions, major releases are costly, infrequent, and difficult to tailor to customer needs, leaving released models with known safety gaps. Unlike full-model fine-tuning or major version updates, our method enables rapid remediation by prepending a compact, learnable prefix to an existing model. This "patch" introduces only 0.003% additional parameters, yet reliably steers model behavior toward that of a safer reference model. Across three critical domains (toxicity mitigation, bias reduction, and harmfulness refusal) policy patches achieve safety improvements comparable to next-generation safety-aligned models while preserving fluency. Our results demonstrate that LLMs can be "patched" much like software, offering vendors and practitioners a practical mechanism for distributing scalable, efficient, and composable safety updates between major model releases.
GP-MoLFormer-Sim: Test Time Molecular Optimization through Contextual Similarity Guidance
Jun 05, 2025Abstract:The ability to design molecules while preserving similarity to a target molecule and/or property is crucial for various applications in drug discovery, chemical design, and biology. We introduce in this paper an efficient training-free method for navigating and sampling from the molecular space with a generative Chemical Language Model (CLM), while using the molecular similarity to the target as a guide. Our method leverages the contextual representations learned from the CLM itself to estimate the molecular similarity, which is then used to adjust the autoregressive sampling strategy of the CLM. At each step of the decoding process, the method tracks the distance of the current generations from the target and updates the logits to encourage the preservation of similarity in generations. We implement the method using a recently proposed $\sim$47M parameter SMILES-based CLM, GP-MoLFormer, and therefore refer to the method as GP-MoLFormer-Sim, which enables a test-time update of the deep generative policy to reflect the contextual similarity to a set of guide molecules. The method is further integrated into a genetic algorithm (GA) and tested on a set of standard molecular optimization benchmarks involving property optimization, molecular rediscovery, and structure-based drug design. Results show that, GP-MoLFormer-Sim, combined with GA (GP-MoLFormer-Sim+GA) outperforms existing training-free baseline methods, when the oracle remains black-box. The findings in this work are a step forward in understanding and guiding the generative mechanisms of CLMs.
Aligning Protein Conformation Ensemble Generation with Physical Feedback
May 30, 2025Abstract:Protein dynamics play a crucial role in protein biological functions and properties, and their traditional study typically relies on time-consuming molecular dynamics (MD) simulations conducted in silico. Recent advances in generative modeling, particularly denoising diffusion models, have enabled efficient accurate protein structure prediction and conformation sampling by learning distributions over crystallographic structures. However, effectively integrating physical supervision into these data-driven approaches remains challenging, as standard energy-based objectives often lead to intractable optimization. In this paper, we introduce Energy-based Alignment (EBA), a method that aligns generative models with feedback from physical models, efficiently calibrating them to appropriately balance conformational states based on their energy differences. Experimental results on the MD ensemble benchmark demonstrate that EBA achieves state-of-the-art performance in generating high-quality protein ensembles. By improving the physical plausibility of generated structures, our approach enhances model predictions and holds promise for applications in structural biology and drug discovery.
PEEL the Layers and Find Yourself: Revisiting Inference-time Data Leakage for Residual Neural Networks
Apr 08, 2025Abstract:This paper explores inference-time data leakage risks of deep neural networks (NNs), where a curious and honest model service provider is interested in retrieving users' private data inputs solely based on the model inference results. Particularly, we revisit residual NNs due to their popularity in computer vision and our hypothesis that residual blocks are a primary cause of data leakage owing to the use of skip connections. By formulating inference-time data leakage as a constrained optimization problem, we propose a novel backward feature inversion method, \textbf{PEEL}, which can effectively recover block-wise input features from the intermediate output of residual NNs. The surprising results in high-quality input data recovery can be explained by the intuition that the output from these residual blocks can be considered as a noisy version of the input and thus the output retains sufficient information for input recovery. We demonstrate the effectiveness of our layer-by-layer feature inversion method on facial image datasets and pre-trained classifiers. Our results show that PEEL outperforms the state-of-the-art recovery methods by an order of magnitude when evaluated by mean squared error (MSE). The code is available at \href{https://github.com/Huzaifa-Arif/PEEL}{https://github.com/Huzaifa-Arif/PEEL}
Fundamental Safety-Capability Trade-offs in Fine-tuning Large Language Models
Mar 24, 2025Abstract:Fine-tuning Large Language Models (LLMs) on some task-specific datasets has been a primary use of LLMs. However, it has been empirically observed that this approach to enhancing capability inevitably compromises safety, a phenomenon also known as the safety-capability trade-off in LLM fine-tuning. This paper presents a theoretical framework for understanding the interplay between safety and capability in two primary safety-aware LLM fine-tuning strategies, providing new insights into the effects of data similarity, context overlap, and alignment loss landscape. Our theoretical results characterize the fundamental limits of the safety-capability trade-off in LLM fine-tuning, which are also validated by numerical experiments.
Can Memory-Augmented Language Models Generalize on Reasoning-in-a-Haystack Tasks?
Mar 10, 2025Abstract:Large language models often expose their brittleness in reasoning tasks, especially while executing long chains of reasoning over context. We propose MemReasoner, a new and simple memory-augmented LLM architecture, in which the memory learns the relative order of facts in context, and enables hopping over them, while the decoder selectively attends to the memory. MemReasoner is trained end-to-end, with optional supporting fact supervision of varying degrees. We train MemReasoner, along with existing memory-augmented transformer models and a state-space model, on two distinct synthetic multi-hop reasoning tasks. Experiments performed under a variety of challenging scenarios, including the presence of long distractor text or target answer changes in test set, show strong generalization of MemReasoner on both single- and two-hop tasks. This generalization of MemReasoner is achieved using none-to-weak supporting fact supervision (using none and 1\% of supporting facts for one- and two-hop tasks, respectively). In contrast, baseline models overall struggle to generalize and benefit far less from using full supporting fact supervision. The results highlight the importance of explicit memory mechanisms, combined with additional weak supervision, for improving large language model's context processing ability toward reasoning tasks.
EpMAN: Episodic Memory AttentioN for Generalizing to Longer Contexts
Feb 20, 2025



Abstract:Recent advances in Large Language Models (LLMs) have yielded impressive successes on many language tasks. However, efficient processing of long contexts using LLMs remains a significant challenge. We introduce \textbf{EpMAN} -- a method for processing long contexts in an \textit{episodic memory} module while \textit{holistically attending to} semantically relevant context chunks. The output of \textit{episodic attention} is then used to reweigh the decoder's self-attention to the stored KV cache of the context during training and generation. When an LLM decoder is trained using \textbf{EpMAN}, its performance on multiple challenging single-hop long-context recall and question-answering benchmarks is found to be stronger and more robust across the range from 16k to 256k tokens than baseline decoders trained with self-attention, and popular retrieval-augmented generation frameworks.
Can Large Language Models Adapt to Other Agents In-Context?
Dec 27, 2024Abstract:As the research community aims to build better AI assistants that are more dynamic and personalized to the diversity of humans that they interact with, there is increased interest in evaluating the theory of mind capabilities of large language models (LLMs). Indeed, several recent studies suggest that LLM theory of mind capabilities are quite impressive, approximating human-level performance. Our paper aims to rebuke this narrative and argues instead that past studies were not directly measuring agent performance, potentially leading to findings that are illusory in nature as a result. We draw a strong distinction between what we call literal theory of mind i.e. measuring the agent's ability to predict the behavior of others and functional theory of mind i.e. adapting to agents in-context based on a rational response to predictions of their behavior. We find that top performing open source LLMs may display strong capabilities in literal theory of mind, depending on how they are prompted, but seem to struggle with functional theory of mind -- even when partner policies are exceedingly simple. Our work serves to highlight the double sided nature of inductive bias in LLMs when adapting to new situations. While this bias can lead to strong performance over limited horizons, it often hinders convergence to optimal long-term behavior.
Combining Domain and Alignment Vectors to Achieve Better Knowledge-Safety Trade-offs in LLMs
Nov 11, 2024



Abstract:There is a growing interest in training domain-expert LLMs that excel in specific technical fields compared to their general-purpose instruction-tuned counterparts. However, these expert models often experience a loss in their safety abilities in the process, making them capable of generating harmful content. As a solution, we introduce an efficient and effective merging-based alignment method called \textsc{MergeAlign} that interpolates the domain and alignment vectors, creating safer domain-specific models while preserving their utility. We apply \textsc{MergeAlign} on Llama3 variants that are experts in medicine and finance, obtaining substantial alignment improvements with minimal to no degradation on domain-specific benchmarks. We study the impact of model merging through model similarity metrics and contributions of individual models being merged. We hope our findings open new research avenues and inspire more efficient development of safe expert LLMs.
SEAL: Safety-enhanced Aligned LLM Fine-tuning via Bilevel Data Selection
Oct 09, 2024
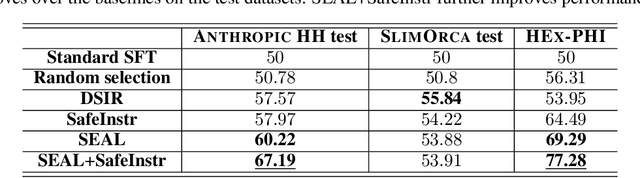

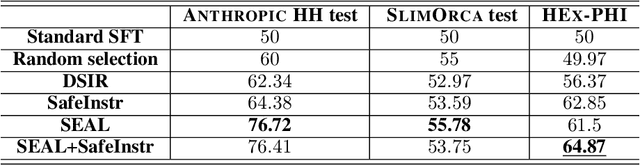
Abstract:Fine-tuning on task-specific data to boost downstream performance is a crucial step for leveraging Large Language Models (LLMs). However, previous studies have demonstrated that fine-tuning the models on several adversarial samples or even benign data can greatly comprise the model's pre-equipped alignment and safety capabilities. In this work, we propose SEAL, a novel framework to enhance safety in LLM fine-tuning. SEAL learns a data ranker based on the bilevel optimization to up rank the safe and high-quality fine-tuning data and down rank the unsafe or low-quality ones. Models trained with SEAL demonstrate superior quality over multiple baselines, with 8.5% and 9.7% win rate increase compared to random selection respectively on Llama-3-8b-Instruct and Merlinite-7b models. Our code is available on github https://github.com/hanshen95/SEAL.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge