Jiarui Lu
COMPASS: A Multi-Turn Benchmark for Tool-Mediated Planning & Preference Optimization
Oct 08, 2025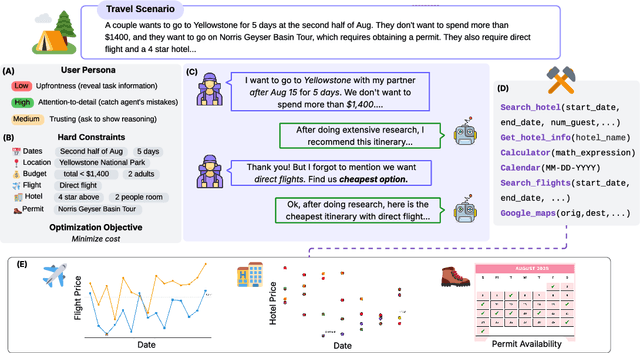
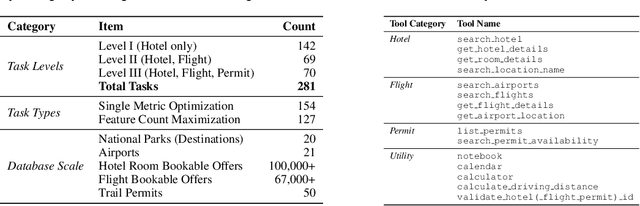
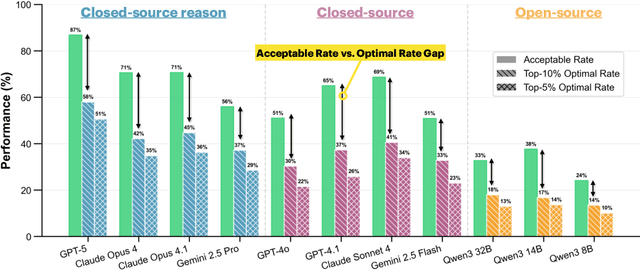
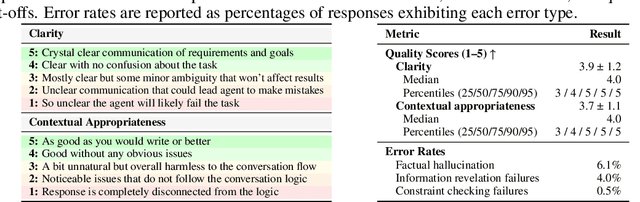
Abstract:Real-world large language model (LLM) agents must master strategic tool use and user preference optimization through multi-turn interactions to assist users with complex planning tasks. We introduce COMPASS (Constrained Optimization through Multi-turn Planning and Strategic Solutions), a benchmark that evaluates agents on realistic travel-planning scenarios. We cast travel planning as a constrained preference optimization problem, where agents must satisfy hard constraints while simultaneously optimizing soft user preferences. To support this, we build a realistic travel database covering transportation, accommodation, and ticketing for 20 U.S. National Parks, along with a comprehensive tool ecosystem that mirrors commercial booking platforms. Evaluating state-of-the-art models, we uncover two critical gaps: (i) an acceptable-optimal gap, where agents reliably meet constraints but fail to optimize preferences, and (ii) a plan-coordination gap, where performance collapses on multi-service (flight and hotel) coordination tasks, especially for open-source models. By grounding reasoning and planning in a practical, user-facing domain, COMPASS provides a benchmark that directly measures an agent's ability to optimize user preferences in realistic tasks, bridging theoretical advances with real-world impact.
Measuring Scientific Capabilities of Language Models with a Systems Biology Dry Lab
Jul 02, 2025
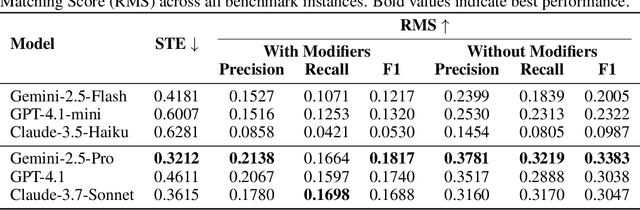
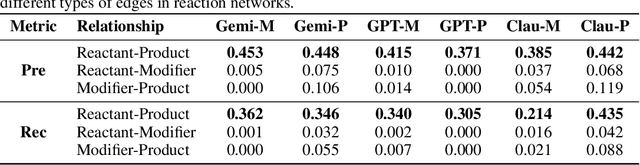
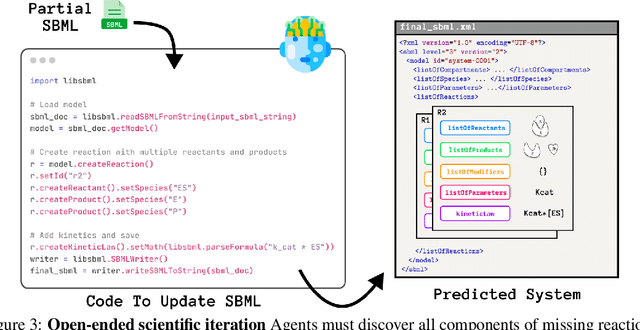
Abstract:Designing experiments and result interpretations are core scientific competencies, particularly in biology, where researchers perturb complex systems to uncover the underlying systems. Recent efforts to evaluate the scientific capabilities of large language models (LLMs) fail to test these competencies because wet-lab experimentation is prohibitively expensive: in expertise, time and equipment. We introduce SciGym, a first-in-class benchmark that assesses LLMs' iterative experiment design and analysis abilities in open-ended scientific discovery tasks. SciGym overcomes the challenge of wet-lab costs by running a dry lab of biological systems. These models, encoded in Systems Biology Markup Language, are efficient for generating simulated data, making them ideal testbeds for experimentation on realistically complex systems. We evaluated six frontier LLMs on 137 small systems, and released a total of 350 systems. Our evaluation shows that while more capable models demonstrated superior performance, all models' performance declined significantly as system complexity increased, suggesting substantial room for improvement in the scientific capabilities of LLM agents.
Aligning Protein Conformation Ensemble Generation with Physical Feedback
May 30, 2025Abstract:Protein dynamics play a crucial role in protein biological functions and properties, and their traditional study typically relies on time-consuming molecular dynamics (MD) simulations conducted in silico. Recent advances in generative modeling, particularly denoising diffusion models, have enabled efficient accurate protein structure prediction and conformation sampling by learning distributions over crystallographic structures. However, effectively integrating physical supervision into these data-driven approaches remains challenging, as standard energy-based objectives often lead to intractable optimization. In this paper, we introduce Energy-based Alignment (EBA), a method that aligns generative models with feedback from physical models, efficiently calibrating them to appropriately balance conformational states based on their energy differences. Experimental results on the MD ensemble benchmark demonstrate that EBA achieves state-of-the-art performance in generating high-quality protein ensembles. By improving the physical plausibility of generated structures, our approach enhances model predictions and holds promise for applications in structural biology and drug discovery.
Self-Evolving Curriculum for LLM Reasoning
May 20, 2025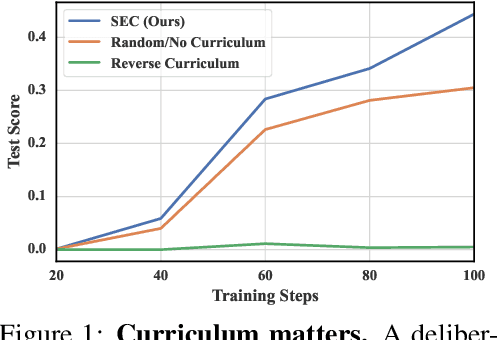
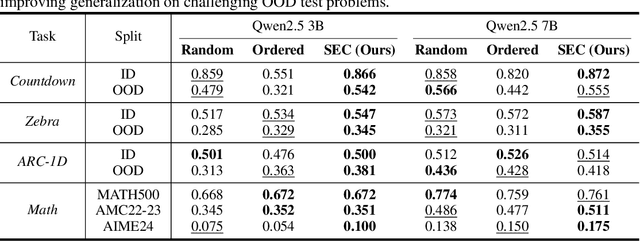
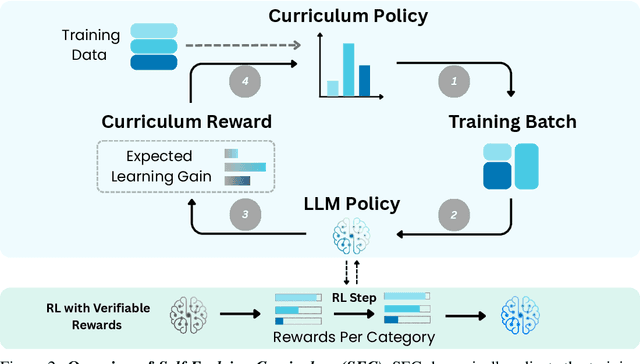
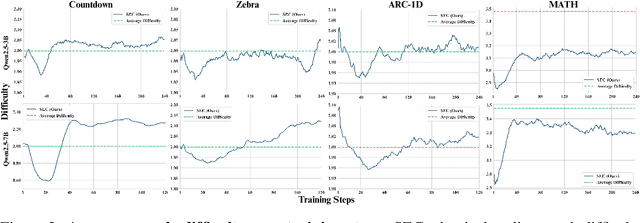
Abstract:Reinforcement learning (RL) has proven effective for fine-tuning large language models (LLMs), significantly enhancing their reasoning abilities in domains such as mathematics and code generation. A crucial factor influencing RL fine-tuning success is the training curriculum: the order in which training problems are presented. While random curricula serve as common baselines, they remain suboptimal; manually designed curricula often rely heavily on heuristics, and online filtering methods can be computationally prohibitive. To address these limitations, we propose Self-Evolving Curriculum (SEC), an automatic curriculum learning method that learns a curriculum policy concurrently with the RL fine-tuning process. Our approach formulates curriculum selection as a non-stationary Multi-Armed Bandit problem, treating each problem category (e.g., difficulty level or problem type) as an individual arm. We leverage the absolute advantage from policy gradient methods as a proxy measure for immediate learning gain. At each training step, the curriculum policy selects categories to maximize this reward signal and is updated using the TD(0) method. Across three distinct reasoning domains: planning, inductive reasoning, and mathematics, our experiments demonstrate that SEC significantly improves models' reasoning capabilities, enabling better generalization to harder, out-of-distribution test problems. Additionally, our approach achieves better skill balance when fine-tuning simultaneously on multiple reasoning domains. These findings highlight SEC as a promising strategy for RL fine-tuning of LLMs.
Reaction-conditioned De Novo Enzyme Design with GENzyme
Nov 10, 2024Abstract:The introduction of models like RFDiffusionAA, AlphaFold3, AlphaProteo, and Chai1 has revolutionized protein structure modeling and interaction prediction, primarily from a binding perspective, focusing on creating ideal lock-and-key models. However, these methods can fall short for enzyme-substrate interactions, where perfect binding models are rare, and induced fit states are more common. To address this, we shift to a functional perspective for enzyme design, where the enzyme function is defined by the reaction it catalyzes. Here, we introduce \textsc{GENzyme}, a \textit{de novo} enzyme design model that takes a catalytic reaction as input and generates the catalytic pocket, full enzyme structure, and enzyme-substrate binding complex. \textsc{GENzyme} is an end-to-end, three-staged model that integrates (1) a catalytic pocket generation and sequence co-design module, (2) a pocket inpainting and enzyme inverse folding module, and (3) a binding and screening module to optimize and predict enzyme-substrate complexes. The entire design process is driven by the catalytic reaction being targeted. This reaction-first approach allows for more accurate and biologically relevant enzyme design, potentially surpassing structure-based and binding-focused models in creating enzymes capable of catalyzing specific reactions. We provide \textsc{GENzyme} code at https://github.com/WillHua127/GENzyme.
Structure Language Models for Protein Conformation Generation
Oct 24, 2024
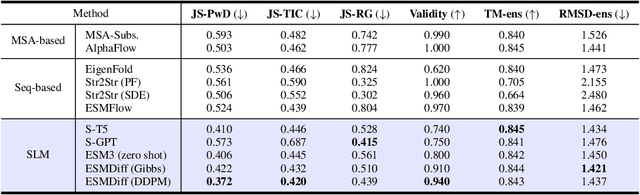
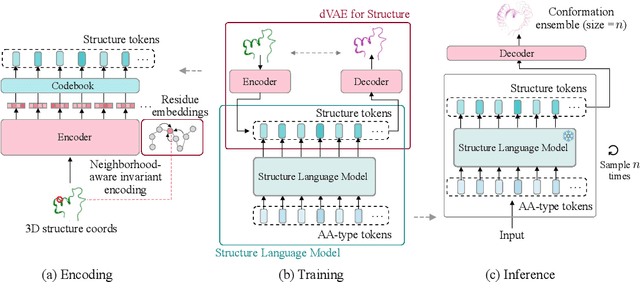
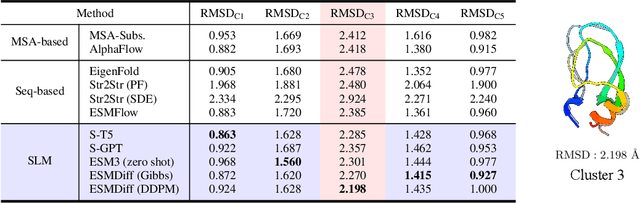
Abstract:Proteins adopt multiple structural conformations to perform their diverse biological functions, and understanding these conformations is crucial for advancing drug discovery. Traditional physics-based simulation methods often struggle with sampling equilibrium conformations and are computationally expensive. Recently, deep generative models have shown promise in generating protein conformations as a more efficient alternative. However, these methods predominantly rely on the diffusion process within a 3D geometric space, which typically centers around the vicinity of metastable states and is often inefficient in terms of runtime. In this paper, we introduce Structure Language Modeling (SLM) as a novel framework for efficient protein conformation generation. Specifically, the protein structures are first encoded into a compact latent space using a discrete variational auto-encoder, followed by conditional language modeling that effectively captures sequence-specific conformation distributions. This enables a more efficient and interpretable exploration of diverse ensemble modes compared to existing methods. Based on this general framework, we instantiate SLM with various popular LM architectures as well as proposing the ESMDiff, a novel BERT-like structure language model fine-tuned from ESM3 with masked diffusion. We verify our approach in various scenarios, including the equilibrium dynamics of BPTI, conformational change pairs, and intrinsically disordered proteins. SLM provides a highly efficient solution, offering a 20-100x speedup than existing methods in generating diverse conformations, shedding light on promising avenues for future research.
ToolSandbox: A Stateful, Conversational, Interactive Evaluation Benchmark for LLM Tool Use Capabilities
Aug 08, 2024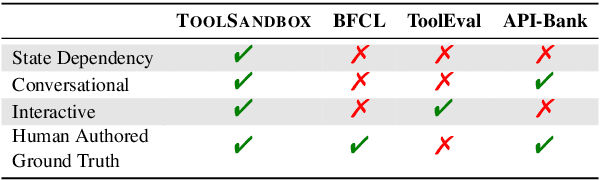
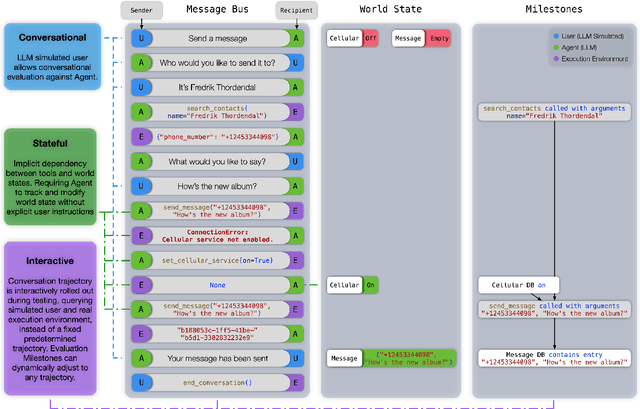
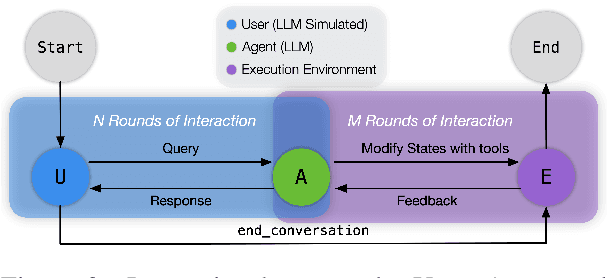
Abstract:Recent large language models (LLMs) advancements sparked a growing research interest in tool assisted LLMs solving real-world challenges, which calls for comprehensive evaluation of tool-use capabilities. While previous works focused on either evaluating over stateless web services (RESTful API), based on a single turn user prompt, or an off-policy dialog trajectory, ToolSandbox includes stateful tool execution, implicit state dependencies between tools, a built-in user simulator supporting on-policy conversational evaluation and a dynamic evaluation strategy for intermediate and final milestones over an arbitrary trajectory. We show that open source and proprietary models have a significant performance gap, and complex tasks like State Dependency, Canonicalization and Insufficient Information defined in ToolSandbox are challenging even the most capable SOTA LLMs, providing brand-new insights into tool-use LLM capabilities. ToolSandbox evaluation framework is released at https://github.com/apple/ToolSandbox
Apple Intelligence Foundation Language Models
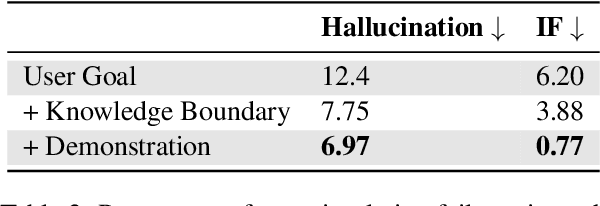
Jul 29, 2024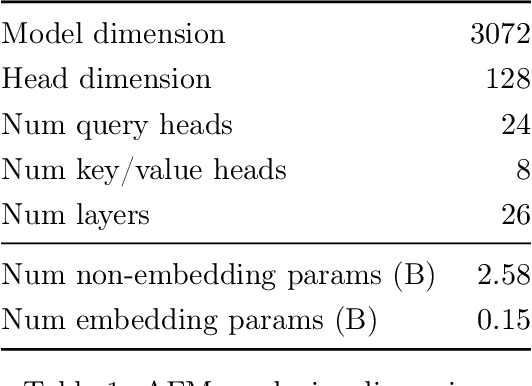
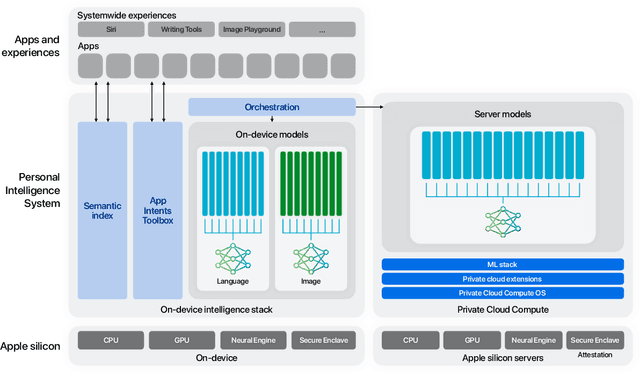


Abstract:We present foundation language models developed to power Apple Intelligence features, including a ~3 billion parameter model designed to run efficiently on devices and a large server-based language model designed for Private Cloud Compute. These models are designed to perform a wide range of tasks efficiently, accurately, and responsibly. This report describes the model architecture, the data used to train the model, the training process, how the models are optimized for inference, and the evaluation results. We highlight our focus on Responsible AI and how the principles are applied throughout the model development.
MMIDR: Teaching Large Language Model to Interpret Multimodal Misinformation via Knowledge Distillation
Mar 21, 2024Abstract:Automatic detection of multimodal misinformation has gained a widespread attention recently. However, the potential of powerful Large Language Models (LLMs) for multimodal misinformation detection remains underexplored. Besides, how to teach LLMs to interpret multimodal misinformation in cost-effective and accessible way is still an open question. To address that, we propose MMIDR, a framework designed to teach LLMs in providing fluent and high-quality textual explanations for their decision-making process of multimodal misinformation. To convert multimodal misinformation into an appropriate instruction-following format, we present a data augmentation perspective and pipeline. This pipeline consists of a visual information processing module and an evidence retrieval module. Subsequently, we prompt the proprietary LLMs with processed contents to extract rationales for interpreting the authenticity of multimodal misinformation. Furthermore, we design an efficient knowledge distillation approach to distill the capability of proprietary LLMs in explaining multimodal misinformation into open-source LLMs. To explore several research questions regarding the performance of LLMs in multimodal misinformation detection tasks, we construct an instruction-following multimodal misinformation dataset and conduct comprehensive experiments. The experimental findings reveal that our MMIDR exhibits sufficient detection performance and possesses the capacity to provide compelling rationales to support its assessments.
Fusing Neural and Physical: Augment Protein Conformation Sampling with Tractable Simulations
Feb 16, 2024Abstract:The protein dynamics are common and important for their biological functions and properties, the study of which usually involves time-consuming molecular dynamics (MD) simulations in silico. Recently, generative models has been leveraged as a surrogate sampler to obtain conformation ensembles with orders of magnitude faster and without requiring any simulation data (a "zero-shot" inference). However, being agnostic of the underlying energy landscape, the accuracy of such generative model may still be limited. In this work, we explore the few-shot setting of such pre-trained generative sampler which incorporates MD simulations in a tractable manner. Specifically, given a target protein of interest, we first acquire some seeding conformations from the pre-trained sampler followed by a number of physical simulations in parallel starting from these seeding samples. Then we fine-tuned the generative model using the simulation trajectories above to become a target-specific sampler. Experimental results demonstrated the superior performance of such few-shot conformation sampler at a tractable computational cost.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge