Roger Trullo
MIPHEI-ViT: Multiplex Immunofluorescence Prediction from H&E Images using ViT Foundation Models
May 15, 2025Abstract:Histopathological analysis is a cornerstone of cancer diagnosis, with Hematoxylin and Eosin (H&E) staining routinely acquired for every patient to visualize cell morphology and tissue architecture. On the other hand, multiplex immunofluorescence (mIF) enables more precise cell type identification via proteomic markers, but has yet to achieve widespread clinical adoption due to cost and logistical constraints. To bridge this gap, we introduce MIPHEI (Multiplex Immunofluorescence Prediction from H&E), a U-Net-inspired architecture that integrates state-of-the-art ViT foundation models as encoders to predict mIF signals from H&E images. MIPHEI targets a comprehensive panel of markers spanning nuclear content, immune lineages (T cells, B cells, myeloid), epithelium, stroma, vasculature, and proliferation. We train our model using the publicly available ORION dataset of restained H&E and mIF images from colorectal cancer tissue, and validate it on two independent datasets. MIPHEI achieves accurate cell-type classification from H&E alone, with F1 scores of 0.88 for Pan-CK, 0.57 for CD3e, 0.56 for SMA, 0.36 for CD68, and 0.30 for CD20, substantially outperforming both a state-of-the-art baseline and a random classifier for most markers. Our results indicate that our model effectively captures the complex relationships between nuclear morphologies in their tissue context, as visible in H&E images and molecular markers defining specific cell types. MIPHEI offers a promising step toward enabling cell-type-aware analysis of large-scale H&E datasets, in view of uncovering relationships between spatial cellular organization and patient outcomes.
RoFormer for Position Aware Multiple Instance Learning in Whole Slide Image Classification
Oct 03, 2023Abstract:Whole slide image (WSI) classification is a critical task in computational pathology. However, the gigapixel-size of such images remains a major challenge for the current state of deep-learning. Current methods rely on multiple-instance learning (MIL) models with frozen feature extractors. Given the the high number of instances in each image, MIL methods have long assumed independence and permutation-invariance of patches, disregarding the tissue structure and correlation between patches. Recent works started studying this correlation between instances but the computational workload of such a high number of tokens remained a limiting factor. In particular, relative position of patches remains unaddressed. We propose to apply a straightforward encoding module, namely a RoFormer layer , relying on memory-efficient exact self-attention and relative positional encoding. This module can perform full self-attention with relative position encoding on patches of large and arbitrary shaped WSIs, solving the need for correlation between instances and spatial modeling of tissues. We demonstrate that our method outperforms state-of-the-art MIL models on three commonly used public datasets (TCGA-NSCLC, BRACS and Camelyon16)) on weakly supervised classification tasks. Code is available at https://github.com/Sanofi-Public/DDS-RoFormerMIL
Image Translation Based Nuclei Segmentation for Immunohistochemistry Images
Aug 12, 2022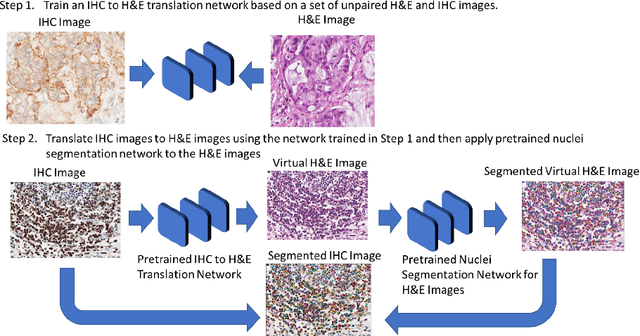
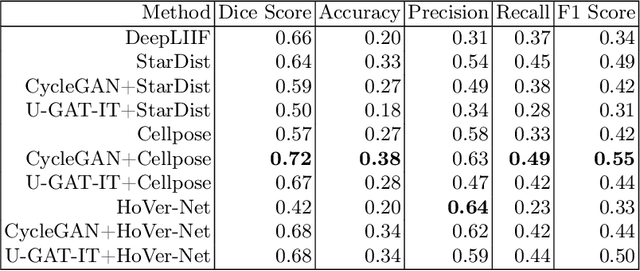
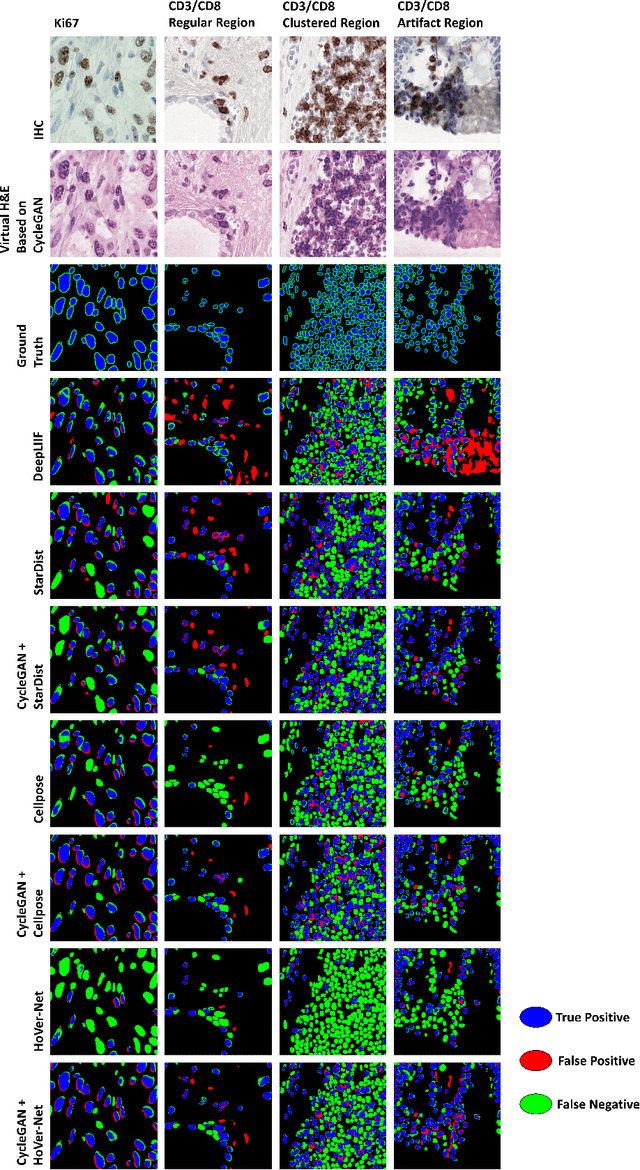
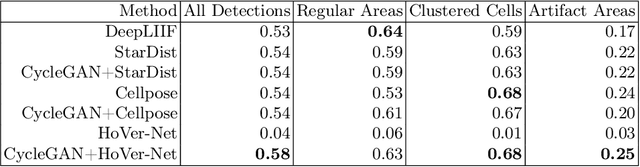
Abstract:Numerous deep learning based methods have been developed for nuclei segmentation for H&E images and have achieved close to human performance. However, direct application of such methods to another modality of images, such as Immunohistochemistry (IHC) images, may not achieve satisfactory performance. Thus, we developed a Generative Adversarial Network (GAN) based approach to translate an IHC image to an H&E image while preserving nuclei location and morphology and then apply pre-trained nuclei segmentation models to the virtual H&E image. We demonstrated that the proposed methods work better than several baseline methods including direct application of state of the art nuclei segmentation methods such as Cellpose and HoVer-Net, trained on H&E and a generative method, DeepLIIF, using two public IHC image datasets.
Descriptive Modeling of Textiles using FE Simulations and Deep Learning
Jun 26, 2021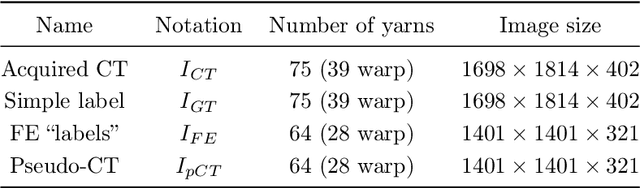
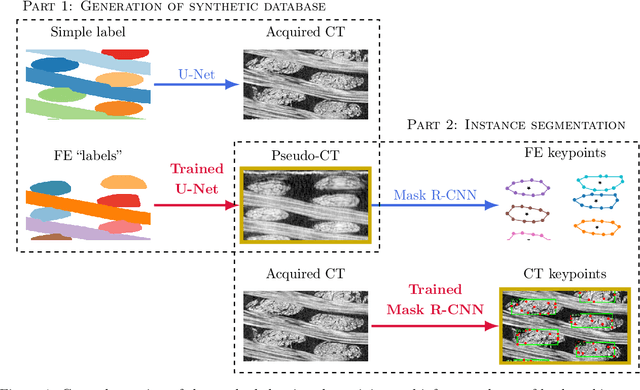
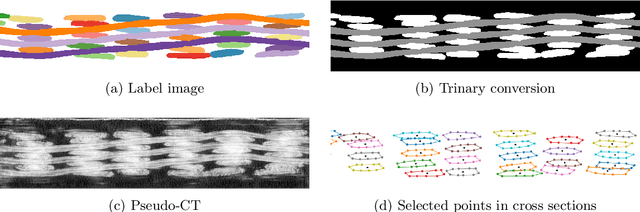
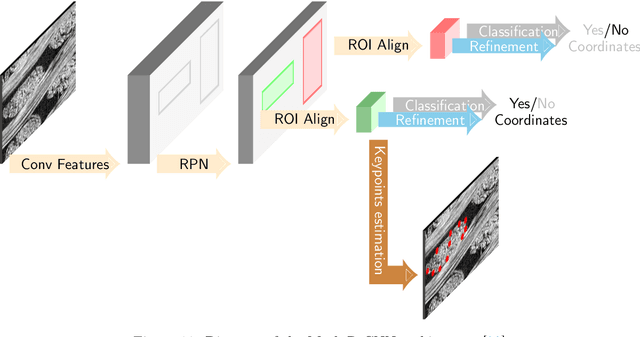
Abstract:In this work we propose a novel and fully automated method for extracting the yarn geometrical features in woven composites so that a direct parametrization of the textile reinforcement is achieved (e.g., FE mesh). Thus, our aim is not only to perform yarn segmentation from tomographic images but rather to provide a complete descriptive modeling of the fabric. As such, this direct approach improves on previous methods that use voxel-wise masks as intermediate representations followed by re-meshing operations (yarn envelope estimation). The proposed approach employs two deep neural network architectures (U-Net and Mask RCNN). First, we train the U-Net to generate synthetic CT images from the corresponding FE simulations. This allows to generate large quantities of annotated data without requiring costly manual annotations. This data is then used to train the Mask R-CNN, which is focused on predicting contour points around each of the yarns in the image. Experimental results show that our method is accurate and robust for performing yarn instance segmentation on CT images, this is further validated by quantitative and qualitative analyses.
Medical Image Synthesis with Context-Aware Generative Adversarial Networks
Dec 16, 2016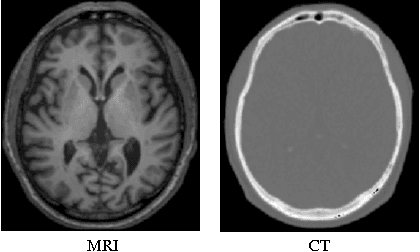
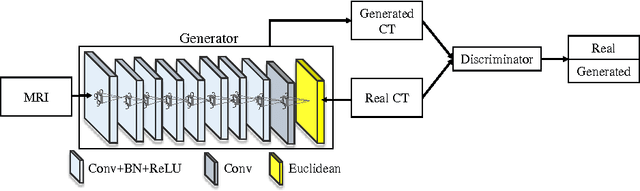
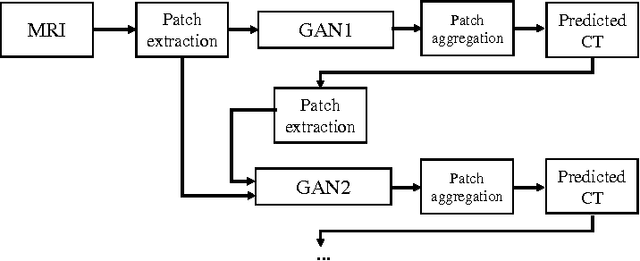
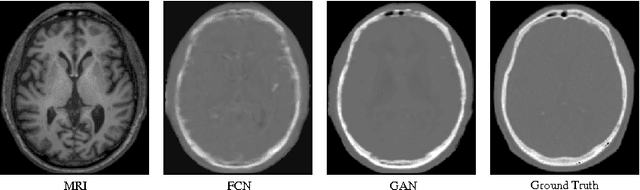
Abstract:Computed tomography (CT) is critical for various clinical applications, e.g., radiotherapy treatment planning and also PET attenuation correction. However, CT exposes radiation during acquisition, which may cause side effects to patients. Compared to CT, magnetic resonance imaging (MRI) is much safer and does not involve any radiations. Therefore, recently, researchers are greatly motivated to estimate CT image from its corresponding MR image of the same subject for the case of radiotherapy planning. In this paper, we propose a data-driven approach to address this challenging problem. Specifically, we train a fully convolutional network to generate CT given an MR image. To better model the nonlinear relationship from MRI to CT and to produce more realistic images, we propose to use the adversarial training strategy and an image gradient difference loss function. We further apply AutoContext Model to implement a context-aware generative adversarial network. Experimental results show that our method is accurate and robust for predicting CT images from MRI images, and also outperforms three state-of-the-art methods under comparison.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge