Melanie Ganz
HQColon: A Hybrid Interactive Machine Learning Pipeline for High Quality Colon Labeling and Segmentation
Feb 28, 2025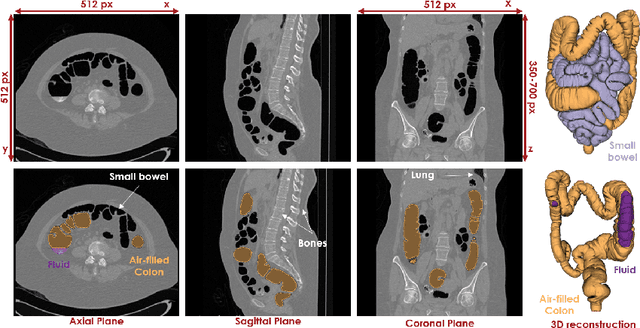
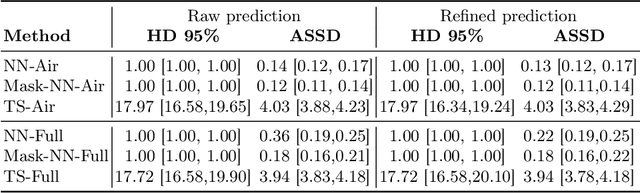
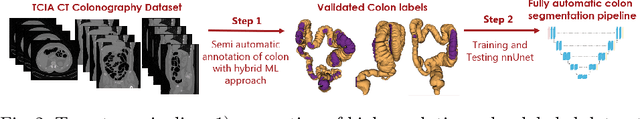
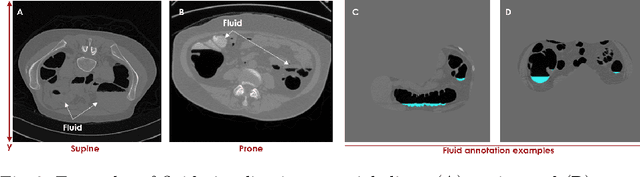
Abstract:High-resolution colon segmentation is crucial for clinical and research applications, such as digital twins and personalized medicine. However, the leading open-source abdominal segmentation tool, TotalSegmentator, struggles with accuracy for the colon, which has a complex and variable shape, requiring time-intensive labeling. Here, we present the first fully automatic high-resolution colon segmentation method. To develop it, we first created a high resolution colon dataset using a pipeline that combines region growing with interactive machine learning to efficiently and accurately label the colon on CT colonography (CTC) images. Based on the generated dataset consisting of 435 labeled CTC images we trained an nnU-Net model for fully automatic colon segmentation. Our fully automatic model achieved an average symmetric surface distance of 0.2 mm (vs. 4.0 mm from TotalSegmentator) and a 95th percentile Hausdorff distance of 1.0 mm (vs. 18 mm from TotalSegmentator). Our segmentation accuracy substantially surpasses TotalSegmentator. We share our trained model and pipeline code, providing the first and only open-source tool for high-resolution colon segmentation. Additionally, we created a large-scale dataset of publicly available high-resolution colon labels.
In the Picture: Medical Imaging Datasets, Artifacts, and their Living Review
Jan 18, 2025


Abstract:Datasets play a critical role in medical imaging research, yet issues such as label quality, shortcuts, and metadata are often overlooked. This lack of attention may harm the generalizability of algorithms and, consequently, negatively impact patient outcomes. While existing medical imaging literature reviews mostly focus on machine learning (ML) methods, with only a few focusing on datasets for specific applications, these reviews remain static -- they are published once and not updated thereafter. This fails to account for emerging evidence, such as biases, shortcuts, and additional annotations that other researchers may contribute after the dataset is published. We refer to these newly discovered findings of datasets as research artifacts. To address this gap, we propose a living review that continuously tracks public datasets and their associated research artifacts across multiple medical imaging applications. Our approach includes a framework for the living review to monitor data documentation artifacts, and an SQL database to visualize the citation relationships between research artifact and dataset. Lastly, we discuss key considerations for creating medical imaging datasets, review best practices for data annotation, discuss the significance of shortcuts and demographic diversity, and emphasize the importance of managing datasets throughout their entire lifecycle. Our demo is publicly available at http://130.226.140.142.
Classification of Radiological Text in Small and Imbalanced Datasets in a Non-English Language
Sep 30, 2024

Abstract:Natural language processing (NLP) in the medical domain can underperform in real-world applications involving small datasets in a non-English language with few labeled samples and imbalanced classes. There is yet no consensus on how to approach this problem. We evaluated a set of NLP models including BERT-like transformers, few-shot learning with sentence transformers (SetFit), and prompted large language models (LLM), using three datasets of radiology reports on magnetic resonance images of epilepsy patients in Danish, a low-resource language. Our results indicate that BERT-like models pretrained in the target domain of radiology reports currently offer the optimal performances for this scenario. Notably, the SetFit and LLM models underperformed compared to BERT-like models, with LLM performing the worst. Importantly, none of the models investigated was sufficiently accurate to allow for text classification without any supervision. However, they show potential for data filtering, which could reduce the amount of manual labeling required.
A sensitivity analysis to quantify the impact of neuroimaging preprocessing strategies on subsequent statistical analyses
Apr 24, 2024Abstract:Even though novel imaging techniques have been successful in studying brain structure and function, the measured biological signals are often contaminated by multiple sources of noise, arising due to e.g. head movements of the individual being scanned, limited spatial/temporal resolution, or other issues specific to each imaging technology. Data preprocessing (e.g. denoising) is therefore critical. Preprocessing pipelines have become increasingly complex over the years, but also more flexible, and this flexibility can have a significant impact on the final results and conclusions of a given study. This large parameter space is often referred to as multiverse analyses. Here, we provide conceptual and practical tools for statistical analyses that can aggregate multiple pipeline results along with a new sensitivity analysis testing for hypotheses across pipelines such as "no effect across all pipelines" or "at least one pipeline with no effect". The proposed framework is generic and can be applied to any multiverse scenario, but we illustrate its use based on positron emission tomography data.
Are demographically invariant models and representations in medical imaging fair?
May 02, 2023

Abstract:Medical imaging models have been shown to encode information about patient demographics (age, race, sex) in their latent representation, raising concerns about their potential for discrimination. Here, we ask whether it is feasible and desirable to train models that do not encode demographic attributes. We consider different types of invariance with respect to demographic attributes - marginal, class-conditional, and counterfactual model invariance - and lay out their equivalence to standard notions of algorithmic fairness. Drawing on existing theory, we find that marginal and class-conditional invariance can be considered overly restrictive approaches for achieving certain fairness notions, resulting in significant predictive performance losses. Concerning counterfactual model invariance, we note that defining medical image counterfactuals with respect to demographic attributes is fraught with complexities. Finally, we posit that demographic encoding may even be considered advantageous if it enables learning a task-specific encoding of demographic features that does not rely on human-constructed categories such as 'race' and 'gender'. We conclude that medical imaging models may need to encode demographic attributes, lending further urgency to calls for comprehensive model fairness assessments in terms of predictive performance.
Why is the winner the best?
Mar 30, 2023
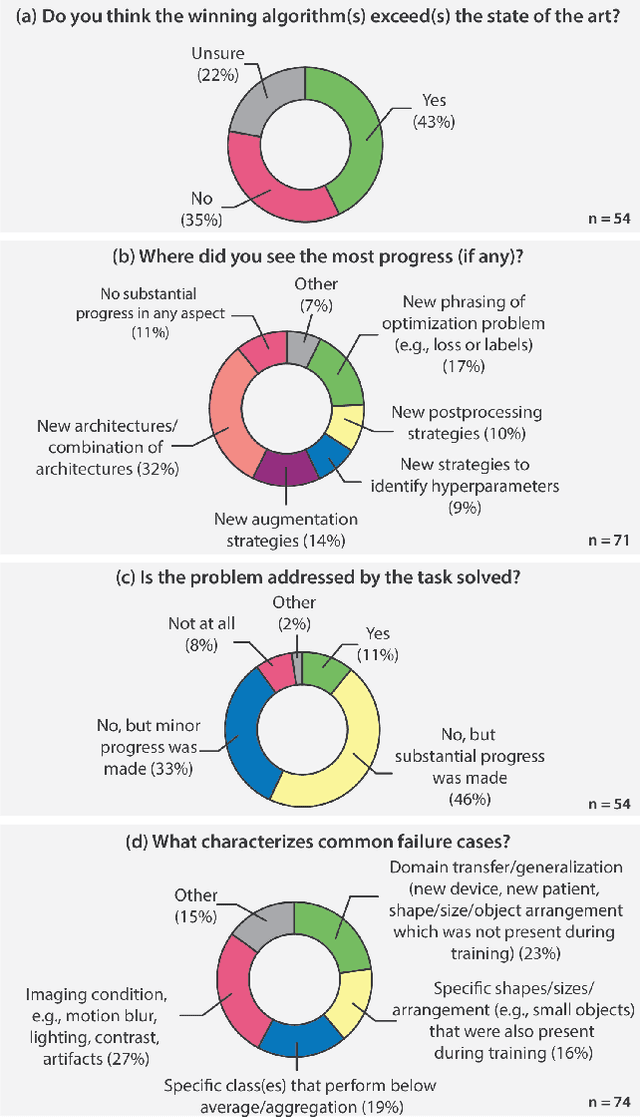
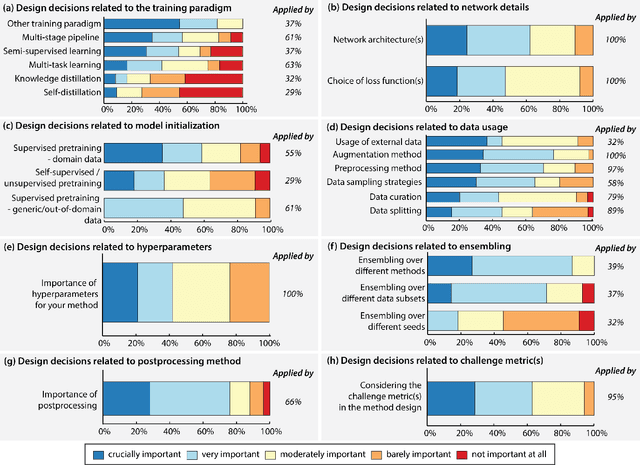
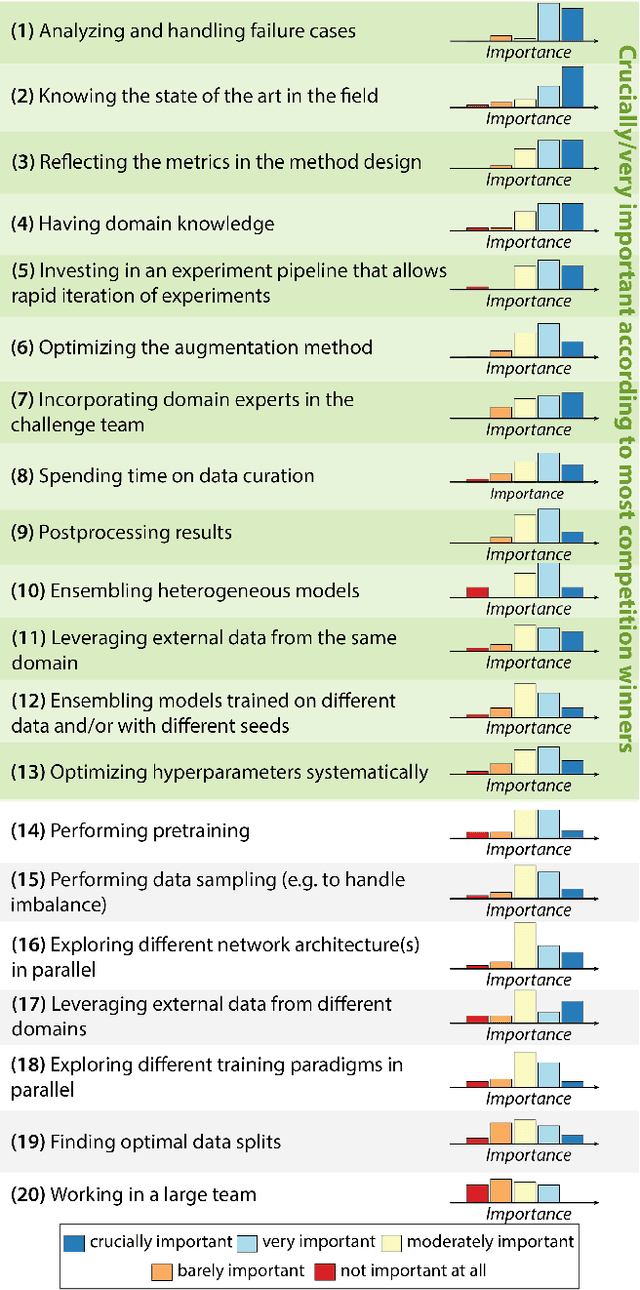
Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
On the fairness of risk score models
Feb 22, 2023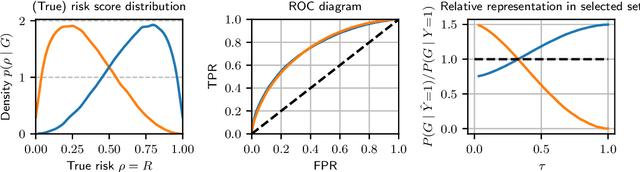
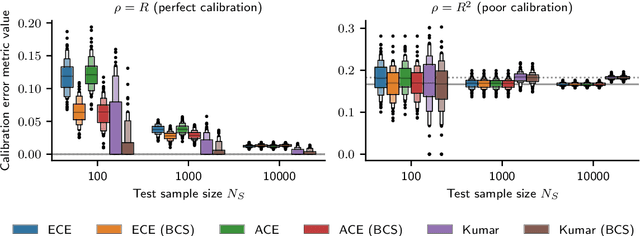
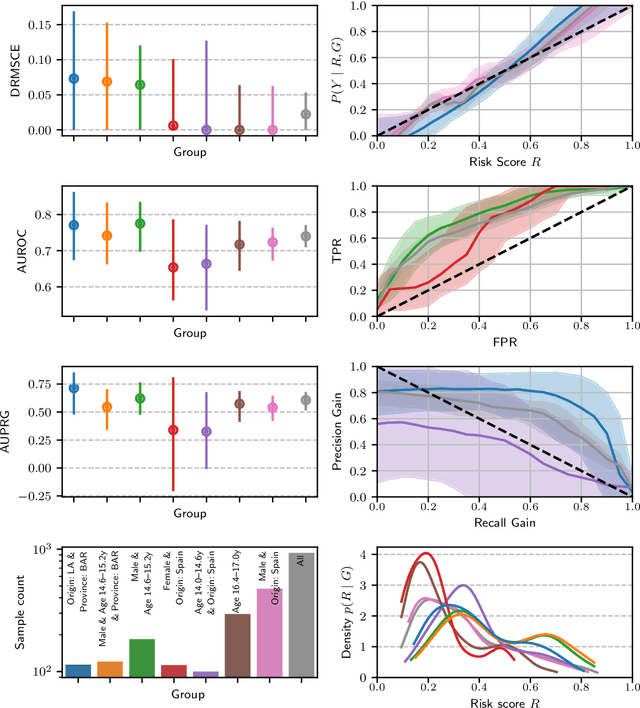
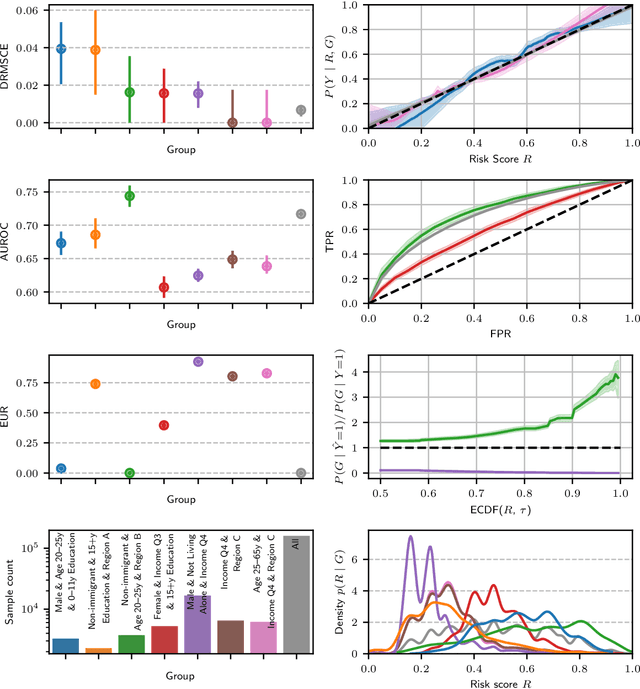
Abstract:Recent work on algorithmic fairness has largely focused on the fairness of discrete decisions, or classifications. While such decisions are often based on risk score models, the fairness of the risk models themselves has received considerably less attention. Risk models are of interest for a number of reasons, including the fact that they communicate uncertainty about the potential outcomes to users, thus representing a way to enable meaningful human oversight. Here, we address fairness desiderata for risk score models. We identify the provision of similar epistemic value to different groups as a key desideratum for risk score fairness. Further, we address how to assess the fairness of risk score models quantitatively, including a discussion of metric choices and meaningful statistical comparisons between groups. In this context, we also introduce a novel calibration error metric that is less sample size-biased than previously proposed metrics, enabling meaningful comparisons between groups of different sizes. We illustrate our methodology - which is widely applicable in many other settings - in two case studies, one in recidivism risk prediction, and one in risk of major depressive disorder (MDD) prediction.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Feature robustness and sex differences in medical imaging: a case study in MRI-based Alzheimer's disease detection
Apr 12, 2022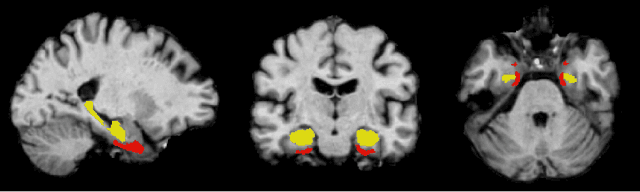

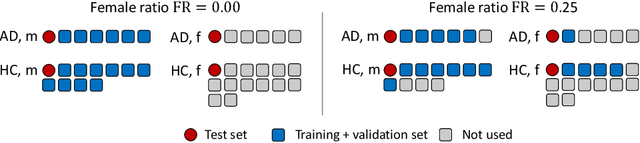
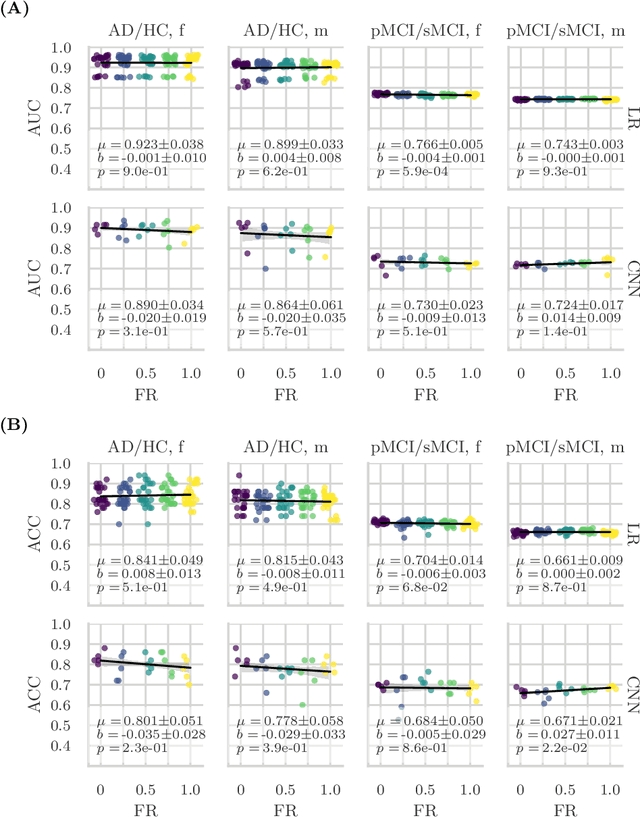
Abstract:Convolutional neural networks have enabled significant improvements in medical image-based disease classification. It has, however, become increasingly clear that these models are susceptible to performance degradation due to spurious correlations and dataset shifts, which may lead to underperformance on underrepresented patient groups, among other problems. In this paper, we compare two classification schemes on the ADNI MRI dataset: a very simple logistic regression model that uses manually selected volumetric features as inputs, and a convolutional neural network trained on 3D MRI data. We assess the robustness of the trained models in the face of varying dataset splits, training set sex composition, and stage of disease. In contrast to earlier work on diagnosing lung diseases based on chest x-ray data, we do not find a strong dependence of model performance for male and female test subjects on the sex composition of the training dataset. Moreover, in our analysis, the low-dimensional model with manually selected features outperforms the 3D CNN, thus emphasizing the need for automatic robust feature extraction methods and the value of manual feature specification (based on prior knowledge) for robustness.
Multi-view consensus CNN for 3D facial landmark placement
Oct 14, 2019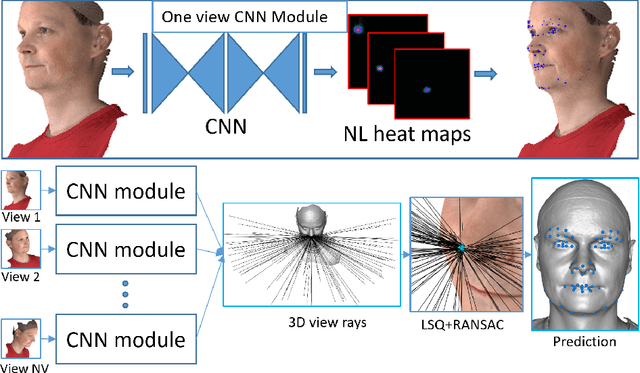
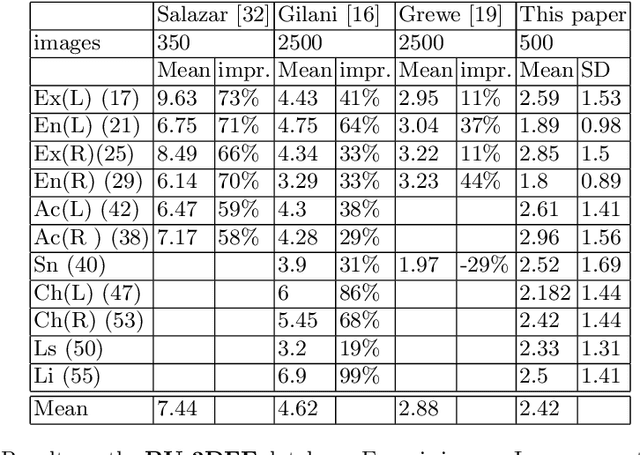
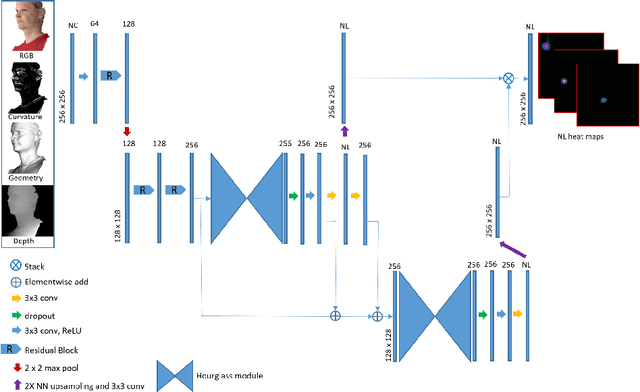
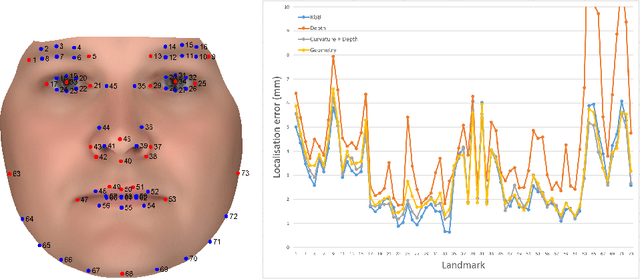
Abstract:The rapid increase in the availability of accurate 3D scanning devices has moved facial recognition and analysis into the 3D domain. 3D facial landmarks are often used as a simple measure of anatomy and it is crucial to have accurate algorithms for automatic landmark placement. The current state-of-the-art approaches have yet to gain from the dramatic increase in performance reported in human pose tracking and 2D facial landmark placement due to the use of deep convolutional neural networks (CNN). Development of deep learning approaches for 3D meshes has given rise to the new subfield called geometric deep learning, where one topic is the adaptation of meshes for the use of deep CNNs. In this work, we demonstrate how methods derived from geometric deep learning, namely multi-view CNNs, can be combined with recent advances in human pose tracking. The method finds 2D landmark estimates and propagates this information to 3D space, where a consensus method determines the accurate 3D face landmark position. We utilise the method on a standard 3D face dataset and show that it outperforms current methods by a large margin. Further, we demonstrate how models trained on 3D range scans can be used to accurately place anatomical landmarks in magnetic resonance images.
* This is a pre-print of an article published in proceedings of the asian conference on computer vision 2018 (LNCS 11361). The final authenticated version is available online at: https://doi.org/10.1007/978-3-030-20887-5_44
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge