Abraham George Smith
HQColon: A Hybrid Interactive Machine Learning Pipeline for High Quality Colon Labeling and Segmentation
Feb 28, 2025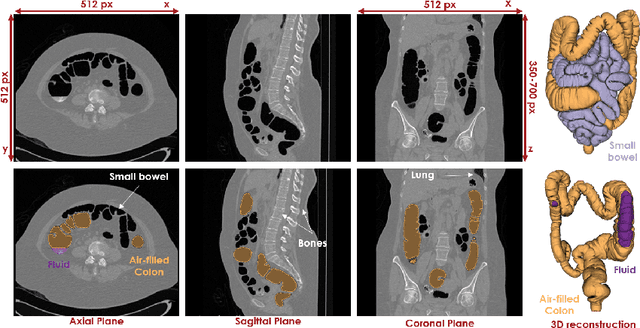
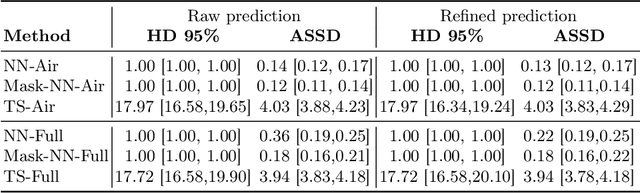
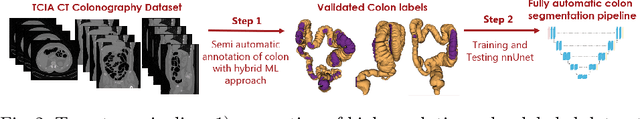
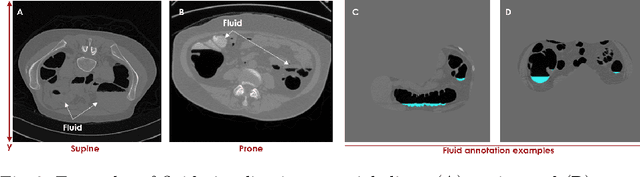
Abstract:High-resolution colon segmentation is crucial for clinical and research applications, such as digital twins and personalized medicine. However, the leading open-source abdominal segmentation tool, TotalSegmentator, struggles with accuracy for the colon, which has a complex and variable shape, requiring time-intensive labeling. Here, we present the first fully automatic high-resolution colon segmentation method. To develop it, we first created a high resolution colon dataset using a pipeline that combines region growing with interactive machine learning to efficiently and accurately label the colon on CT colonography (CTC) images. Based on the generated dataset consisting of 435 labeled CTC images we trained an nnU-Net model for fully automatic colon segmentation. Our fully automatic model achieved an average symmetric surface distance of 0.2 mm (vs. 4.0 mm from TotalSegmentator) and a 95th percentile Hausdorff distance of 1.0 mm (vs. 18 mm from TotalSegmentator). Our segmentation accuracy substantially surpasses TotalSegmentator. We share our trained model and pipeline code, providing the first and only open-source tool for high-resolution colon segmentation. Additionally, we created a large-scale dataset of publicly available high-resolution colon labels.
Prediction of post-radiotherapy recurrence volumes in head and neck squamous cell carcinoma using 3D U-Net segmentation
Aug 16, 2023



Abstract:Locoregional recurrences (LRR) are still a frequent site of treatment failure for head and neck squamous cell carcinoma (HNSCC) patients. Identification of high risk subvolumes based on pretreatment imaging is key to biologically targeted radiation therapy. We investigated the extent to which a Convolutional neural network (CNN) is able to predict LRR volumes based on pre-treatment 18F-fluorodeoxyglucose positron emission tomography (FDG-PET)/computed tomography (CT) scans in HNSCC patients and thus the potential to identify biological high risk volumes using CNNs. For 37 patients who had undergone primary radiotherapy for oropharyngeal squamous cell carcinoma, five oncologists contoured the relapse volumes on recurrence CT scans. Datasets of pre-treatment FDG-PET/CT, gross tumour volume (GTV) and contoured relapse for each of the patients were randomly divided into training (n=23), validation (n=7) and test (n=7) datasets. We compared a CNN trained from scratch, a pre-trained CNN, a SUVmax threshold approach, and using the GTV directly. The SUVmax threshold method included 5 out of the 7 relapse origin points within a volume of median 4.6 cubic centimetres (cc). Both the GTV contour and best CNN segmentations included the relapse origin 6 out of 7 times with median volumes of 28 and 18 cc respectively. The CNN included the same or greater number of relapse volume POs, with significantly smaller relapse volumes. Our novel findings indicate that CNNs may predict LRR, yet further work on dataset development is required to attain clinically useful prediction accuracy.
Localise to segment: crop to improve organ at risk segmentation accuracy
Apr 10, 2023Abstract:Increased organ at risk segmentation accuracy is required to reduce cost and complications for patients receiving radiotherapy treatment. Some deep learning methods for the segmentation of organs at risk use a two stage process where a localisation network first crops an image to the relevant region and then a locally specialised network segments the cropped organ of interest. We investigate the accuracy improvements brought about by such a localisation stage by comparing to a single-stage baseline network trained on full resolution images. We find that localisation approaches can improve both training time and stability and a two stage process involving both a localisation and organ segmentation network provides a significant increase in segmentation accuracy for the spleen, pancreas and heart from the Medical Segmentation Decathlon dataset. We also observe increased benefits of localisation for smaller organs. Source code that recreates the main results is available at \href{https://github.com/Abe404/localise_to_segment}{this https URL}.
RootPainter3D: Interactive-machine-learning enables rapid and accurate contouring for radiotherapy
Jun 22, 2021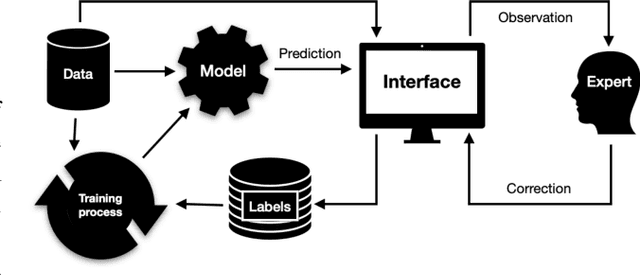
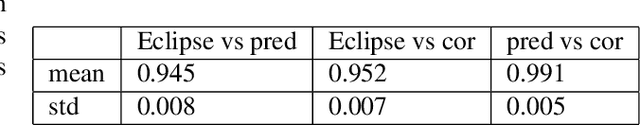
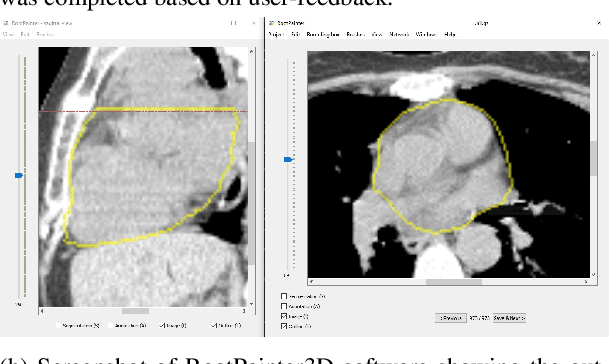
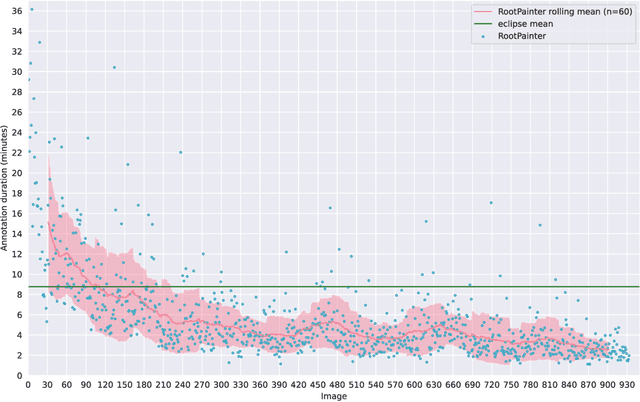
Abstract:Organ-at-risk contouring is still a bottleneck in radiotherapy, with many deep learning methods falling short of promised results when evaluated on clinical data. We investigate the accuracy and time-savings resulting from the use of an interactive-machine-learning method for an organ-at-risk contouring task. We compare the method to the Eclipse contouring software and find strong agreement with manual delineations, with a dice score of 0.95. The annotations created using corrective-annotation also take less time to create as more images are annotated, resulting in substantial time savings compared to manual methods, with hearts that take 2 minutes and 2 seconds to delineate on average, after 923 images have been delineated, compared to 7 minutes and 1 seconds when delineating manually. Our experiment demonstrates that interactive-machine-learning with corrective-annotation provides a fast and accessible way for non computer-scientists to train deep-learning models to segment their own structures of interest as part of routine clinical workflows. Source code is available at \href{https://github.com/Abe404/RootPainter3D}{this HTTPS URL}.
Segmentation of Roots in Soil with U-Net
Mar 18, 2019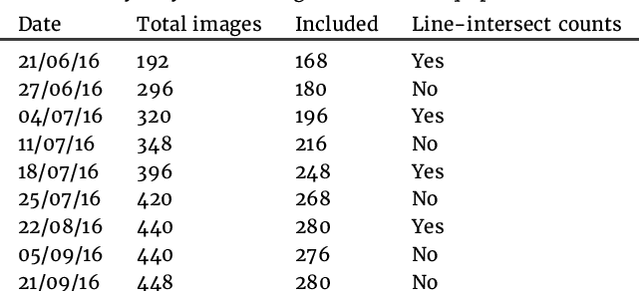

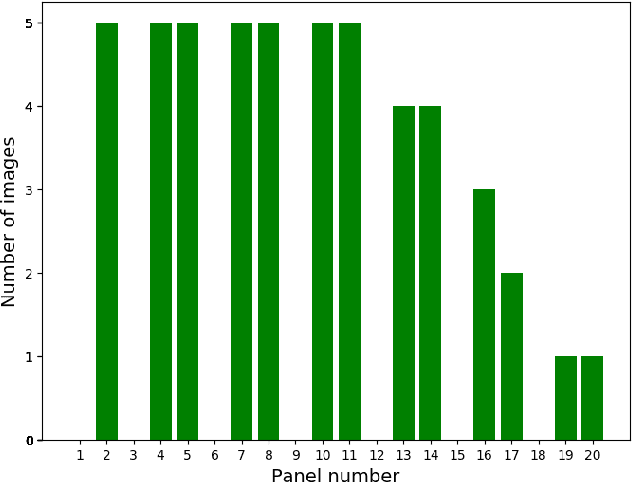

Abstract:Plant root research can provide a way to attain stress-tolerant crops that produce greater yield in a diverse array of conditions. Phenotyping roots in soil is often challenging due to the roots being difficult to access and the use of time consuming manual methods. Rhizotrons allow visual inspection of root growth through transparent surfaces. Agronomists currently manually label photographs of roots obtained from rhizotrons using a line-intersect method to obtain root length density and rooting depth measurements which are essential for their experiments. We investigate the effectiveness of an automated image segmentation method based on the U-Net Convolutional Neural Network (CNN) architecture to enable such measurements. We design a data-set of 50 annotated Chicory (Cichorium intybus L.) root images which we use to train, validate and test the system and compare against a baseline built using the Frangi vesselness filter. We obtain metrics using manual annotations and line-intersect counts. Our results on the held out data show our proposed automated segmentation system to be a viable solution for detecting and quantifying roots. We evaluate our system using 867 images for which we have obtained line-intersect counts, attaining a Spearman rank correlation of 0.9748 and an $r^2$ of 0.9217. We also achieve an $F_1$ of 0.7 when comparing the automated segmentation to the manual annotations, with our automated segmentation system producing segmentations with higher quality than the manual annotations for large portions of the image.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge