Ivan Richter Vogelius
Localise to segment: crop to improve organ at risk segmentation accuracy
Apr 10, 2023Abstract:Increased organ at risk segmentation accuracy is required to reduce cost and complications for patients receiving radiotherapy treatment. Some deep learning methods for the segmentation of organs at risk use a two stage process where a localisation network first crops an image to the relevant region and then a locally specialised network segments the cropped organ of interest. We investigate the accuracy improvements brought about by such a localisation stage by comparing to a single-stage baseline network trained on full resolution images. We find that localisation approaches can improve both training time and stability and a two stage process involving both a localisation and organ segmentation network provides a significant increase in segmentation accuracy for the spleen, pancreas and heart from the Medical Segmentation Decathlon dataset. We also observe increased benefits of localisation for smaller organs. Source code that recreates the main results is available at \href{https://github.com/Abe404/localise_to_segment}{this https URL}.
Explicit Temporal Embedding in Deep Generative Latent Models for Longitudinal Medical Image Synthesis
Jan 13, 2023



Abstract:Medical imaging plays a vital role in modern diagnostics and treatment. The temporal nature of disease or treatment progression often results in longitudinal data. Due to the cost and potential harm, acquiring large medical datasets necessary for deep learning can be difficult. Medical image synthesis could help mitigate this problem. However, until now, the availability of GANs capable of synthesizing longitudinal volumetric data has been limited. To address this, we use the recent advances in latent space-based image editing to propose a novel joint learning scheme to explicitly embed temporal dependencies in the latent space of GANs. This, in contrast to previous methods, allows us to synthesize continuous, smooth, and high-quality longitudinal volumetric data with limited supervision. We show the effectiveness of our approach on three datasets containing different longitudinal dependencies. Namely, modeling a simple image transformation, breathing motion, and tumor regression, all while showing minimal disentanglement. The implementation is made available online at https://github.com/julschoen/Temp-GAN.
RootPainter3D: Interactive-machine-learning enables rapid and accurate contouring for radiotherapy
Jun 22, 2021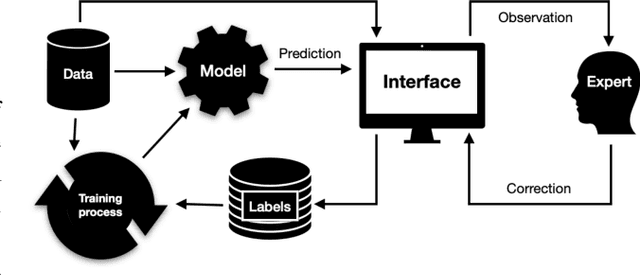
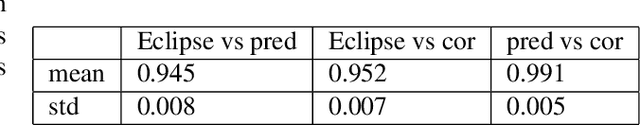
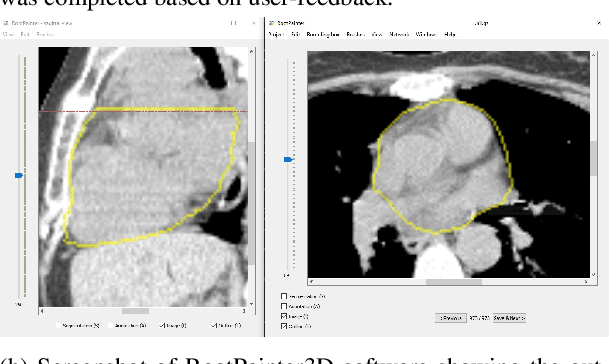
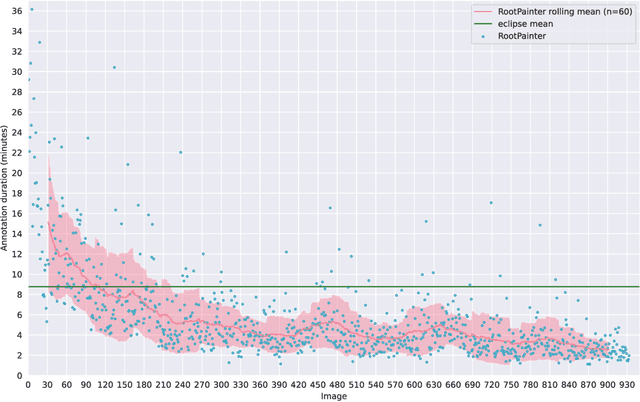
Abstract:Organ-at-risk contouring is still a bottleneck in radiotherapy, with many deep learning methods falling short of promised results when evaluated on clinical data. We investigate the accuracy and time-savings resulting from the use of an interactive-machine-learning method for an organ-at-risk contouring task. We compare the method to the Eclipse contouring software and find strong agreement with manual delineations, with a dice score of 0.95. The annotations created using corrective-annotation also take less time to create as more images are annotated, resulting in substantial time savings compared to manual methods, with hearts that take 2 minutes and 2 seconds to delineate on average, after 923 images have been delineated, compared to 7 minutes and 1 seconds when delineating manually. Our experiment demonstrates that interactive-machine-learning with corrective-annotation provides a fast and accessible way for non computer-scientists to train deep-learning models to segment their own structures of interest as part of routine clinical workflows. Source code is available at \href{https://github.com/Abe404/RootPainter3D}{this HTTPS URL}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge