Lena Specht
Prediction of post-radiotherapy recurrence volumes in head and neck squamous cell carcinoma using 3D U-Net segmentation
Aug 16, 2023



Abstract:Locoregional recurrences (LRR) are still a frequent site of treatment failure for head and neck squamous cell carcinoma (HNSCC) patients. Identification of high risk subvolumes based on pretreatment imaging is key to biologically targeted radiation therapy. We investigated the extent to which a Convolutional neural network (CNN) is able to predict LRR volumes based on pre-treatment 18F-fluorodeoxyglucose positron emission tomography (FDG-PET)/computed tomography (CT) scans in HNSCC patients and thus the potential to identify biological high risk volumes using CNNs. For 37 patients who had undergone primary radiotherapy for oropharyngeal squamous cell carcinoma, five oncologists contoured the relapse volumes on recurrence CT scans. Datasets of pre-treatment FDG-PET/CT, gross tumour volume (GTV) and contoured relapse for each of the patients were randomly divided into training (n=23), validation (n=7) and test (n=7) datasets. We compared a CNN trained from scratch, a pre-trained CNN, a SUVmax threshold approach, and using the GTV directly. The SUVmax threshold method included 5 out of the 7 relapse origin points within a volume of median 4.6 cubic centimetres (cc). Both the GTV contour and best CNN segmentations included the relapse origin 6 out of 7 times with median volumes of 28 and 18 cc respectively. The CNN included the same or greater number of relapse volume POs, with significantly smaller relapse volumes. Our novel findings indicate that CNNs may predict LRR, yet further work on dataset development is required to attain clinically useful prediction accuracy.
RootPainter3D: Interactive-machine-learning enables rapid and accurate contouring for radiotherapy
Jun 22, 2021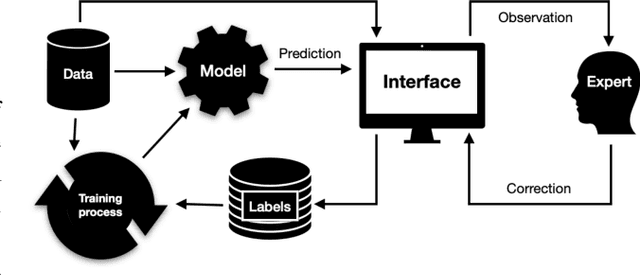
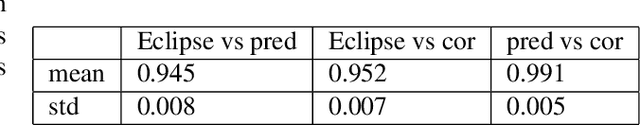
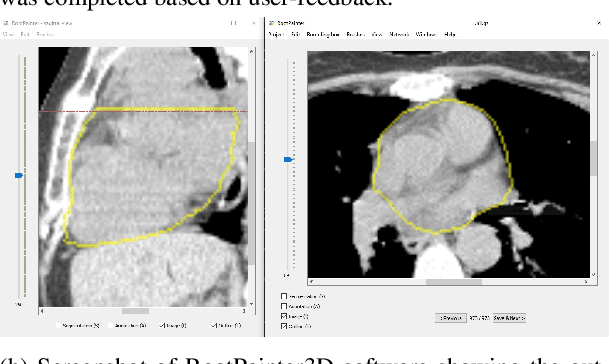
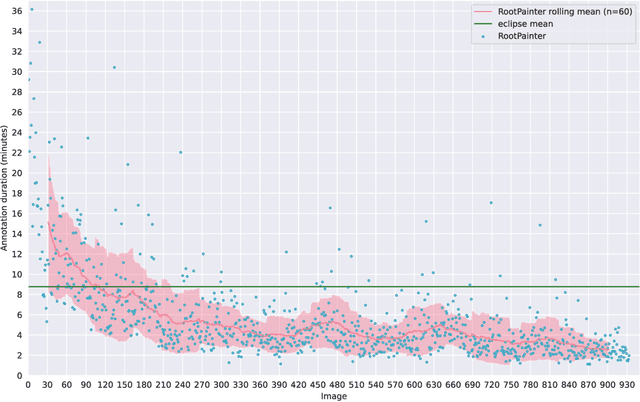
Abstract:Organ-at-risk contouring is still a bottleneck in radiotherapy, with many deep learning methods falling short of promised results when evaluated on clinical data. We investigate the accuracy and time-savings resulting from the use of an interactive-machine-learning method for an organ-at-risk contouring task. We compare the method to the Eclipse contouring software and find strong agreement with manual delineations, with a dice score of 0.95. The annotations created using corrective-annotation also take less time to create as more images are annotated, resulting in substantial time savings compared to manual methods, with hearts that take 2 minutes and 2 seconds to delineate on average, after 923 images have been delineated, compared to 7 minutes and 1 seconds when delineating manually. Our experiment demonstrates that interactive-machine-learning with corrective-annotation provides a fast and accessible way for non computer-scientists to train deep-learning models to segment their own structures of interest as part of routine clinical workflows. Source code is available at \href{https://github.com/Abe404/RootPainter3D}{this HTTPS URL}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge