Kenny Erleben
MERA: Multimodal and Multiscale Self-Explanatory Model with Considerably Reduced Annotation for Lung Nodule Diagnosis
Apr 27, 2025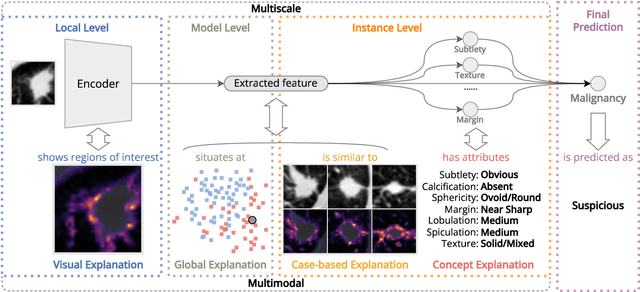
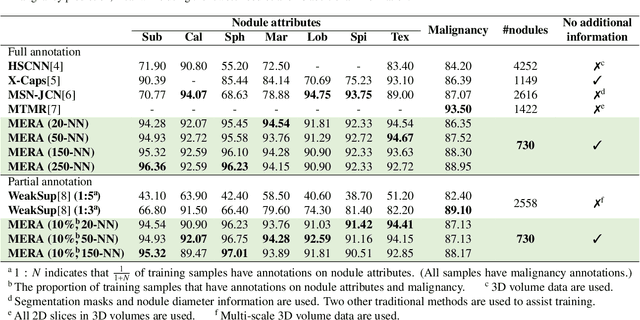
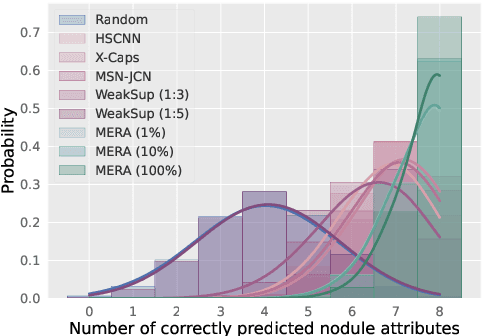
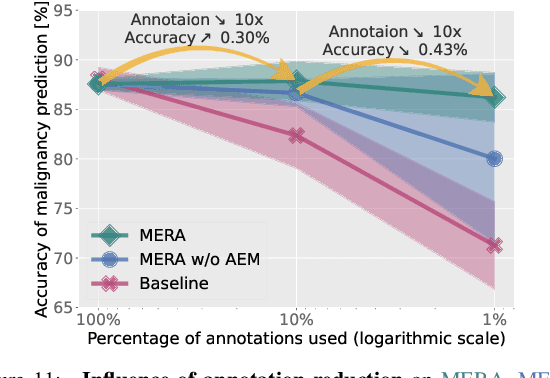
Abstract:Lung cancer, a leading cause of cancer-related deaths globally, emphasises the importance of early detection for better patient outcomes. Pulmonary nodules, often early indicators of lung cancer, necessitate accurate, timely diagnosis. Despite Explainable Artificial Intelligence (XAI) advances, many existing systems struggle providing clear, comprehensive explanations, especially with limited labelled data. This study introduces MERA, a Multimodal and Multiscale self-Explanatory model designed for lung nodule diagnosis with considerably Reduced Annotation requirements. MERA integrates unsupervised and weakly supervised learning strategies (self-supervised learning techniques and Vision Transformer architecture for unsupervised feature extraction) and a hierarchical prediction mechanism leveraging sparse annotations via semi-supervised active learning in the learned latent space. MERA explains its decisions on multiple levels: model-level global explanations via semantic latent space clustering, instance-level case-based explanations showing similar instances, local visual explanations via attention maps, and concept explanations using critical nodule attributes. Evaluations on the public LIDC dataset show MERA's superior diagnostic accuracy and self-explainability. With only 1% annotated samples, MERA achieves diagnostic accuracy comparable to or exceeding state-of-the-art methods requiring full annotation. The model's inherent design delivers comprehensive, robust, multilevel explanations aligned closely with clinical practice, enhancing trustworthiness and transparency. Demonstrated viability of unsupervised and weakly supervised learning lowers the barrier to deploying diagnostic AI in broader medical domains. Our complete code is open-source available: https://github.com/diku-dk/credanno.
Neural Kinematic Bases for Fluids
Apr 22, 2025



Abstract:We propose mesh-free fluid simulations that exploit a kinematic neural basis for velocity fields represented by an MLP. We design a set of losses that ensures that these neural bases satisfy fundamental physical properties such as orthogonality, divergence-free, boundary alignment, and smoothness. Our neural bases can then be used to fit an input sketch of a flow, which will inherit the same fundamental properties from the bases. We then can animate such flow in real-time using standard time integrators. Our neural bases can accommodate different domains and naturally extend to three dimensions.
HQColon: A Hybrid Interactive Machine Learning Pipeline for High Quality Colon Labeling and Segmentation
Feb 28, 2025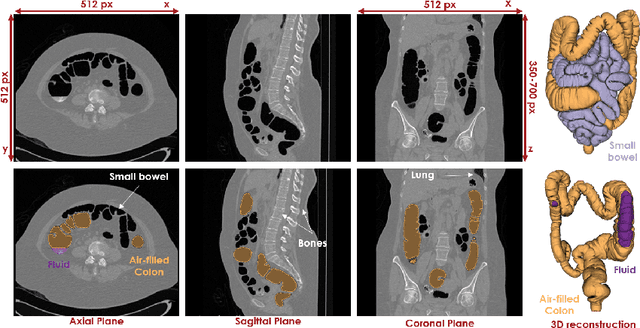
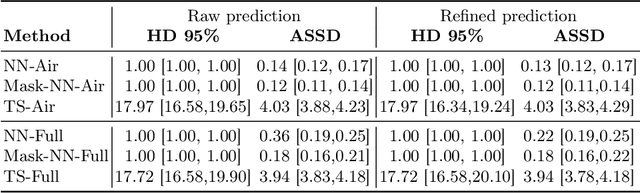
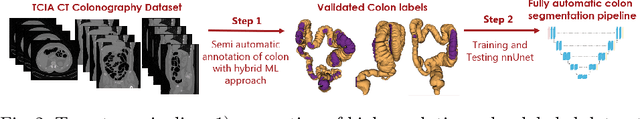
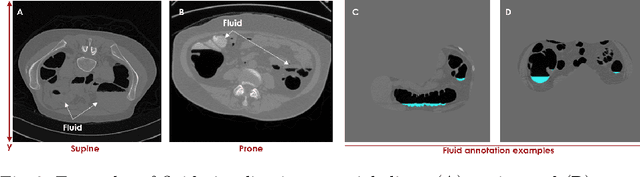
Abstract:High-resolution colon segmentation is crucial for clinical and research applications, such as digital twins and personalized medicine. However, the leading open-source abdominal segmentation tool, TotalSegmentator, struggles with accuracy for the colon, which has a complex and variable shape, requiring time-intensive labeling. Here, we present the first fully automatic high-resolution colon segmentation method. To develop it, we first created a high resolution colon dataset using a pipeline that combines region growing with interactive machine learning to efficiently and accurately label the colon on CT colonography (CTC) images. Based on the generated dataset consisting of 435 labeled CTC images we trained an nnU-Net model for fully automatic colon segmentation. Our fully automatic model achieved an average symmetric surface distance of 0.2 mm (vs. 4.0 mm from TotalSegmentator) and a 95th percentile Hausdorff distance of 1.0 mm (vs. 18 mm from TotalSegmentator). Our segmentation accuracy substantially surpasses TotalSegmentator. We share our trained model and pipeline code, providing the first and only open-source tool for high-resolution colon segmentation. Additionally, we created a large-scale dataset of publicly available high-resolution colon labels.
Differentiable Rendering as a Way to Program Cable-Driven Soft Robots
Apr 11, 2024


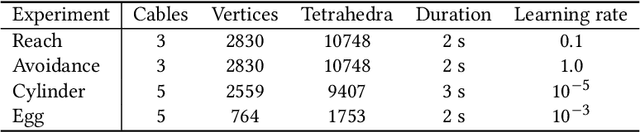
Abstract:Soft robots have gained increased popularity in recent years due to their adaptability and compliance. In this paper, we use a digital twin model of cable-driven soft robots to learn control parameters in simulation. In doing so, we take advantage of differentiable rendering as a way to instruct robots to complete tasks such as point reach, gripping an object, and obstacle avoidance. This approach simplifies the mathematical description of such complicated tasks and removes the need for landmark points and their tracking. Our experiments demonstrate the applicability of our method.
Extremely weakly-supervised blood vessel segmentation with physiologically based synthesis and domain adaptation
May 26, 2023



Abstract:Accurate analysis and modeling of renal functions require a precise segmentation of the renal blood vessels. Micro-CT scans provide image data at higher resolutions, making more small vessels near the renal cortex visible. Although deep-learning-based methods have shown state-of-the-art performance in automatic blood vessel segmentations, they require a large amount of labeled training data. However, voxel-wise labeling in micro-CT scans is extremely time-consuming given the huge volume sizes. To mitigate the problem, we simulate synthetic renal vascular trees physiologically while generating corresponding scans of the simulated trees by training a generative model on unlabeled scans. This enables the generative model to learn the mapping implicitly without the need for explicit functions to emulate the image acquisition process. We further propose an additional segmentation branch over the generative model trained on the generated scans. We demonstrate that the model can directly segment blood vessels on real scans and validate our method on both 3D micro-CT scans of rat kidneys and a proof-of-concept experiment on 2D retinal images. Code and 3D results are available at https://github.com/miccai2023anony/RenalVesselSeg
Deep Learning-Assisted Localisation of Nanoparticles in synthetically generated two-photon microscopy images
Mar 17, 2023Abstract:Tracking single molecules is instrumental for quantifying the transport of molecules and nanoparticles in biological samples, e.g., in brain drug delivery studies. Existing intensity-based localisation methods are not developed for imaging with a scanning microscope, typically used for in vivo imaging. Low signal-to-noise ratios, movement of molecules out-of-focus, and high motion blur on images recorded with scanning two-photon microscopy (2PM) in vivo pose a challenge to the accurate localisation of molecules. Using data-driven models is challenging due to low data volumes, typical for in vivo experiments. We developed a 2PM image simulator to supplement scarce training data. The simulator mimics realistic motion blur, background fluorescence, and shot noise observed in vivo imaging. Training a data-driven model with simulated data improves localisation quality in simulated images and shows why intensity-based methods fail.
A Hybrid Approach to Full-Scale Reconstruction of Renal Arterial Network
Mar 03, 2023Abstract:The renal vasculature, acting as a resource distribution network, plays an important role in both the physiology and pathophysiology of the kidney. However, no imaging techniques allow an assessment of the structure and function of the renal vasculature due to limited spatial and temporal resolution. To develop realistic computer simulations of renal function, and to develop new image-based diagnostic methods based on artificial intelligence, it is necessary to have a realistic full-scale model of the renal vasculature. We propose a hybrid framework to build subject-specific models of the renal vascular network by using semi-automated segmentation of large arteries and estimation of cortex area from a micro-CT scan as a starting point, and by adopting the Global Constructive Optimization algorithm for generating smaller vessels. Our results show a statistical correspondence between the reconstructed data and existing anatomical data obtained from a rat kidney with respect to morphometric and hemodynamic parameters.
cRedAnno+: Annotation Exploitation in Self-Explanatory Lung Nodule Diagnosis
Nov 07, 2022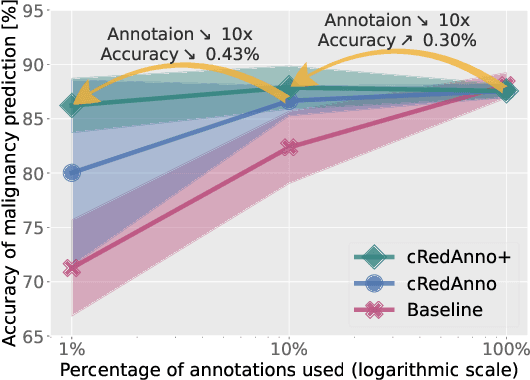
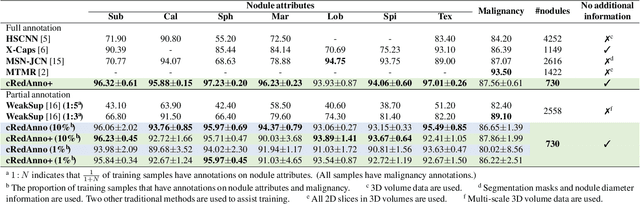
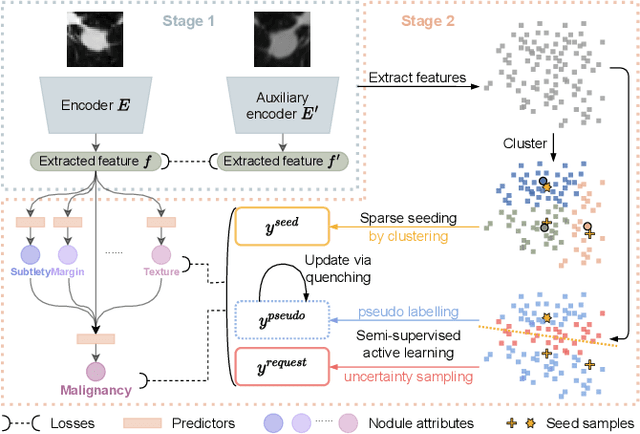
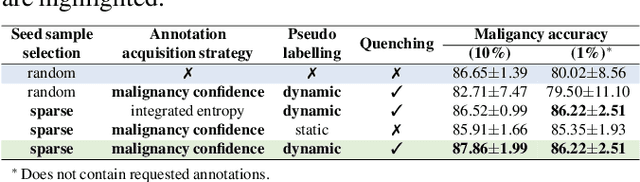
Abstract:Recently, attempts have been made to reduce annotation requirements in feature-based self-explanatory models for lung nodule diagnosis. As a representative, cRedAnno achieves competitive performance with considerably reduced annotation needs by introducing self-supervised contrastive learning to do unsupervised feature extraction. However, it exhibits unstable performance under scarce annotation conditions. To improve the accuracy and robustness of cRedAnno, we propose an annotation exploitation mechanism by conducting semi-supervised active learning with sparse seeding and training quenching in the learned semantically meaningful reasoning space to jointly utilise the extracted features, annotations, and unlabelled data. The proposed approach achieves comparable or even higher malignancy prediction accuracy with 10x fewer annotations, meanwhile showing better robustness and nodule attribute prediction accuracy under the condition of 1% annotations. Our complete code is open-source available: https://github.com/diku-dk/credanno.
Auto-segmentation of Hip Joints using MultiPlanar UNet with Transfer learning
Aug 18, 2022
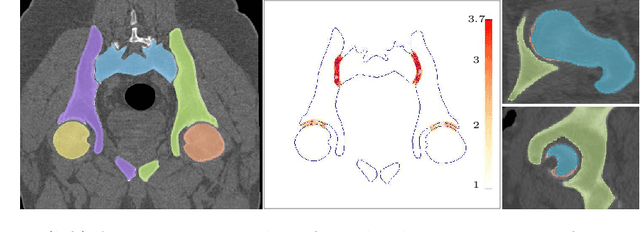


Abstract:Accurate geometry representation is essential in developing finite element models. Although generally good, deep-learning segmentation approaches with only few data have difficulties in accurately segmenting fine features, e.g., gaps and thin structures. Subsequently, segmented geometries need labor-intensive manual modifications to reach a quality where they can be used for simulation purposes. We propose a strategy that uses transfer learning to reuse datasets with poor segmentation combined with an interactive learning step where fine-tuning of the data results in anatomically accurate segmentations suitable for simulations. We use a modified MultiPlanar UNet that is pre-trained using inferior hip joint segmentation combined with a dedicated loss function to learn the gap regions and post-processing to correct tiny inaccuracies on symmetric classes due to rotational invariance. We demonstrate this robust yet conceptually simple approach applied with clinically validated results on publicly available computed tomography scans of hip joints. Code and resulting 3D models are available at: https://github.com/MICCAI2022-155/AuToSeg}
Reducing Annotation Need in Self-Explanatory Models for Lung Nodule Diagnosis
Jun 27, 2022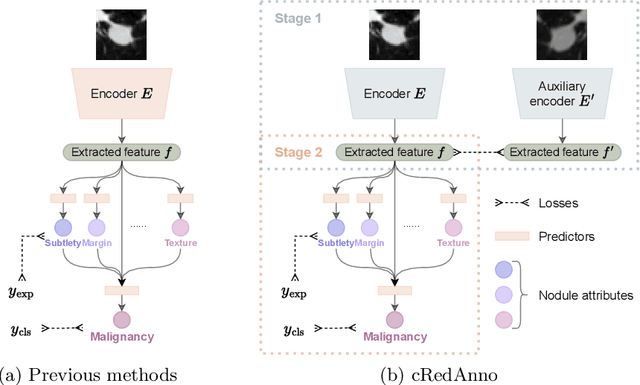
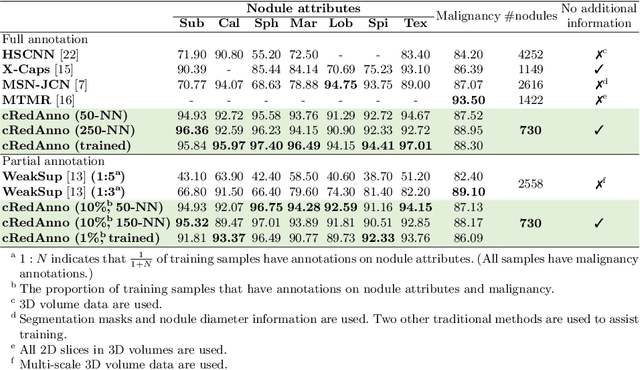
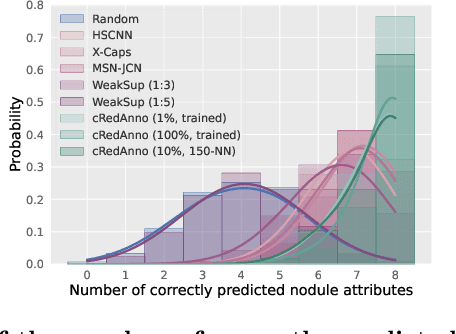
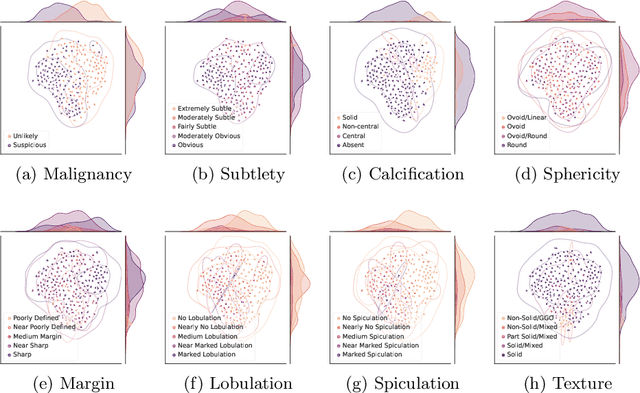
Abstract:Feature-based self-explanatory methods explain their classification in terms of human-understandable features. In the medical imaging community, this semantic matching of clinical knowledge adds significantly to the trustworthiness of the AI. However, the cost of additional annotation of features remains a pressing issue. We address this problem by proposing cRedAnno, a data-/annotation-efficient self-explanatory approach for lung nodule diagnosis. cRedAnno considerably reduces the annotation need by introducing self-supervised contrastive learning to alleviate the burden of learning most parameters from annotation, replacing end-to-end training with two-stage training. When training with hundreds of nodule samples and only 1% of their annotations, cRedAnno achieves competitive accuracy in predicting malignancy, meanwhile significantly surpassing most previous works in predicting nodule attributes. Visualisation of the learned space further indicates that the correlation between the clustering of malignancy and nodule attributes coincides with clinical knowledge. Our complete code is open-source available: https://github.com/ludles/credanno.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge